Dear afni,
AFNI version info (afni -ver): Jan 11 2024 (Version AFNI_24.0.01 'Caracalla')
I'm using SSwarper and afni_proc for data preprocessing, and I've collected some warnings from the program's output after processing the data of eight subjects. There are some warnings that I am confused and hope to get some advice.
I've listed all the output warnings, and the ones I think can be ignored are marked by . Others I have questions about are marked as . I've saved all the output, for more details please tell me. Only one T1w and one epi data for each subject.
SSwarper
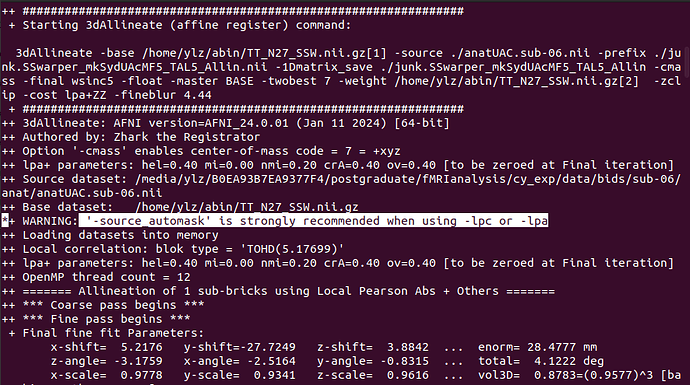
- When SSwarper calls the 3dAllineate program, such a warning is given. When using the -lpc or -lpa option, -source_automask is recommended, but SSwarper doesn't seem to offer this option. Perhaps it can or can only be ignored.
*+ WARNING: '-source_automask' is strongly recommended when using -lpc or -lpa
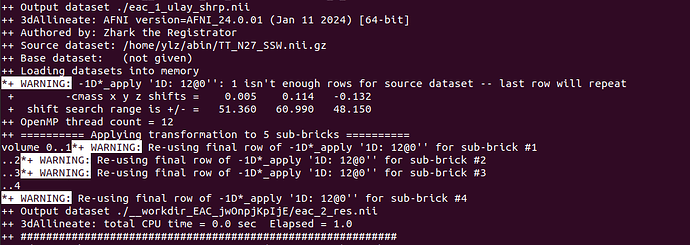
- It's still a 3dAllineate program output, why does it output a warning of "isn't enough rows for source dataset", is it feasible to repeat the last row of data? What caused the absence?
The warning for "Applying transformation to 5 sub-bricks" is output because the same transformation is applied. So it can be ignored,right?
*+ WARNING: -1D*_apply '1D: 12@0'': 1 isn't enough rows for source dataset -- last row will repeat
*+ WARNING: Re-using final row of -1D*_apply '1D: 12@0'' for sub-brick
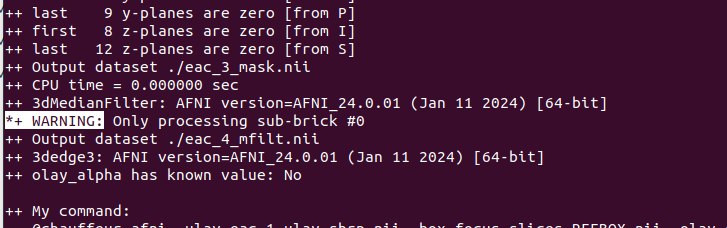
- Output by 3dMedianFilter. Can be ignored?
*+ WARNING: Only processing sub-brick #0
afni_proc.py
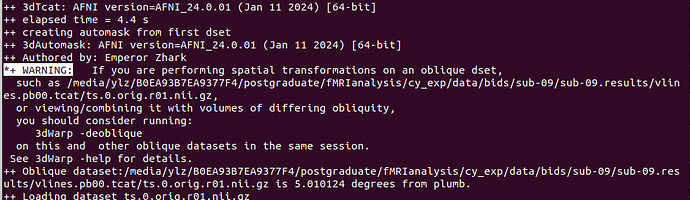
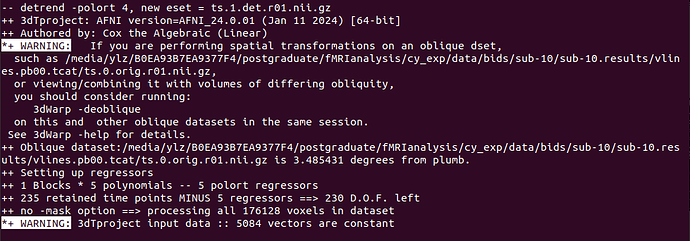
- A oblique warning, I know afni_proc handles it, so this warning can be ignored! I'm not worried.
*+ WARNING: If you are performing spatial transformations on an oblique dset
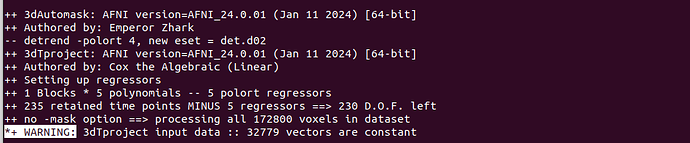
- I read the post below and there are a small number of voxels that are constant which could be considered to be parts out of the brain, but in my case the constant about 18%, is there something wrong? How do I check my data? (32779/172800≈18.97% ). This warning came up several times, ranging from 5,000+ to 30,000+ voxels.
warning in 3dTproject: input data :: 18 vectors are constant
*+ WARNING: 3dTproject input data :: 32779 vectors are constant
- Some other warnings. It looks like it could be ignored, but I'm not sure.
*+ WARNING: No mask being used? That *could* be OK, but thought I'd let you know...
*+ WARNING: input 'a' is not used in the expression
- This warning can be ignored! See below.
afni proc warning "Input dataset is not 3D+time; assuming TR=1.0
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
- This is a warning output by 3dAllineate, and I don't know what the warning is trying to tell me.
*+ WARNING: No output dataset will be calculated
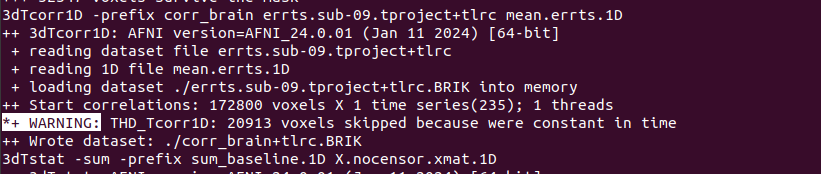
- This may be a warning output by afni_proc when performing regression? Same as the 3dTproject, is it unusual for so many voxels are constants.
THD_Tcorr1D: 20913 voxels skipped because were constant in time
- Can it be ignored as well as the 7th?
*+ WARNING: Input dataset has 1 sub-brick ==> -tdiff is turned off
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
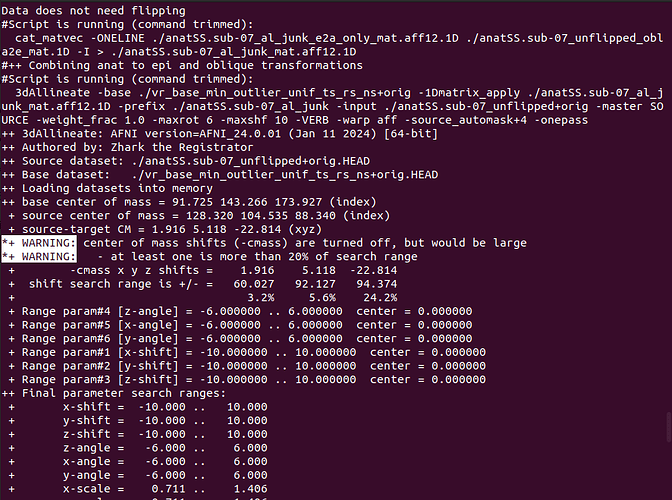
- I don't know much about this, does not use the center-of-mass calculation to determine an initial shift cause bad results? Or how to check if the results generated without cmass are good?
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
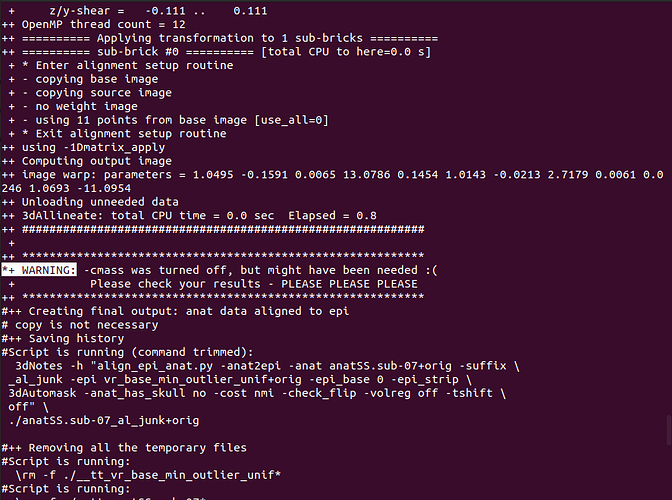
- Same as the previous one
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
QC report
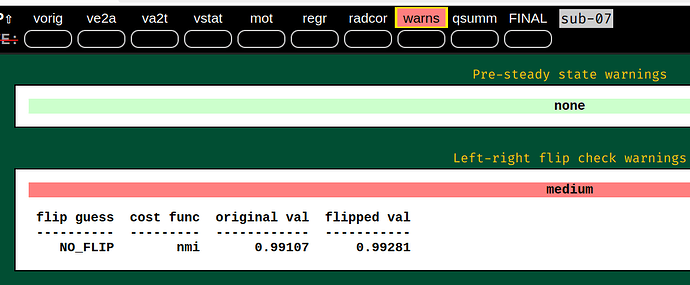
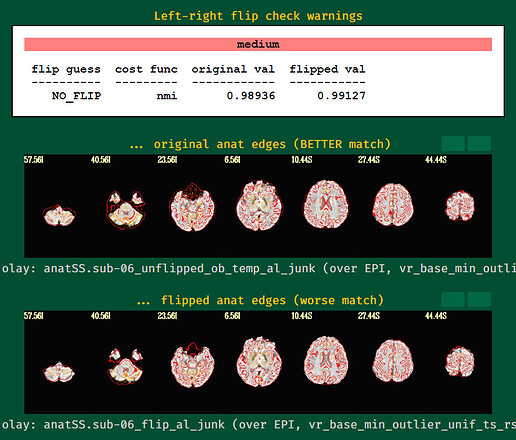
- The quality control visualization looks great, but there's a warning, is it trying to express that the results are better after flipped?
Scripts
@SSwarper \
-input ${subject}_T1w.nii \
-base $SSW_template \
-subid ${subject}
afni_proc.py \
-subj_id $subject \
-dsets func/${subject}_bold.nii \
-copy_anat anat/anatSS.${subject}.nii \
-blocks despike tshift align tlrc volreg blur \
mask scale regress \
-tcat_remove_first_trs $dummy_scans \
-align_unifize_epi local \
-anat_has_skull no \
-align_opts_aea -cost nmi \
-check_flip \
-tlrc_base $template \
-tlrc_NL_warp \
-tlrc_NL_warped_dsets anat/anatQQ.${subject}.nii \
anat/anatQQ.${subject}.aff12.1D \
anat/anatQQ.${subject}_WARP.nii \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-blur_size 4 \
-mask_epi_anat yes \
-regress_censor_motion 0.2 \
-regress_censor_outliers 0.05 \
-regress_bandpass 0.01 0.1 \
-regress_apply_mot_types deriv \
-regress_run_clustsim no \
-regress_est_blur_epits \
-regress_est_blur_errts \
-html_review_style pythonic \
-execute
So many questions, any reply would be appreciated!
Thanks,
Yang
Hi, Yang-
There are some intermediate warnings that can pop up in the processing stream. Many of these will probably not mean that something bad is necessarily happening, just that if the final outputs have problems then these might be some things to consider. But it is also great to ask about them to check.
One thing to note first: as of Jan 20, 2024 (so, about 9 days after your current version here was built), we added sswarper2 to the distribution. It is an updated version of @SSwarper, with essentially the same syntax of use, and I would recommend updating your code and using sswarper2.
Note: I think you should add -prefix .. to either your @SSwarper or sswarper2 command to specify the output filename.
To your warning list:
-
I think all intermediate processing warnings 1-12 can be ignored, likely. How do your alignment outputs look? The ones related to alignment probably only apply if the initial alignments between datasets are very off and a good alignment doesn't converge.
-
To the QC report: this one has an associated image that it would be important to see. It relates to this potential issue of left-right flipping between a subject's EPI and anatomical:
Glen DR, Taylor PA, Buchsbaum BR, Cox RW, Reynolds RC (2020). Beware (Surprisingly Common) Left-Right Flips in Your MRI Data: An Efficient and Robust Method to Check MRI Dataset Consistency Using AFNI. Front. Neuroinformatics 14.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7263312/
I'm surprised to see the EPI-anatomical cost functions be so similar between "as is" and "flipped". It would be good to see the image there. NB: see the next comment, about cost functions.
-
In your afni_proc.py command, I see you are using the nmi cost function. Is there a reason you are not using the more typically recommended lpc+ZZ? That is what we used for discerning left-right flips for the most part. It tends to be a more powerful cost function for EPI-anatomical alignment, as long as their is enough EPI tissue contrast visible.
-
More generally about your afni_proc.py command, perhaps you might want to see this recent draft and related code to some processing choices (GitHub link therein). For example, I might consider adding these opts:
# Add a post-volreg TSNR plot to the APQC HTML:
-volreg_compute_tsnr yes
and
# Explicitly specify the voxel size of the final EPI data:
-volreg_warp_dxyz ${final_dxyz}
-
Are you sure you want to bandpass the resting state date? It has downsides to consider, even though it is widely done in many papers. Consider this Resting State Note.
-
Is there a reason you don't want to include demean, which would normally be done, in this option:
-regress_apply_mot_types demean deriv
?
-
Your blur size is 4. We typically would recommend blurring single echo data by a factor of 1.5-2 times the minimum voxel dimension of the input EPI. What is your input voxel size?
--pt
Hi ptaylor,
Thank you for your reply!
Glad to hear about the @SSwarper update, I'll update my afni and use the latest sswarper2!
Here are some additions:
@SSwarper works fine, anat to template alignment is good! But EPI to anat alignment isn't as good as anat to template.- About cost fucntions, my EPI tissue don't have enough contrast, so when I use lpc+ZZ, the EPI to anat alignment is bad.
- Thanks for the suggestion about resting-state preprocessing! I'd consider dropping the bandpass filtering on my data. And add demean to the -regress_apply_mot_types option.
- The EPI data with a voxel size of 3mm, all output data are voxels of 3mm size.
Supplementary images(take one subject as an example):
-
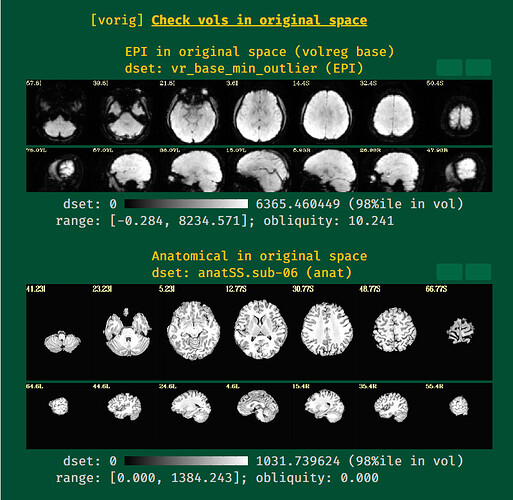
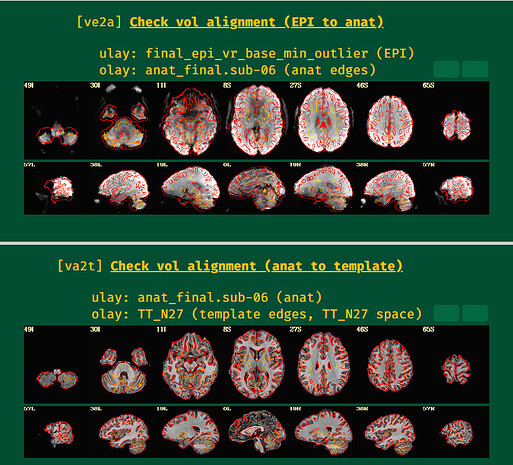
initial EPI and T1w images
-
ve2a and va2t images
-
left-right flipping images
-
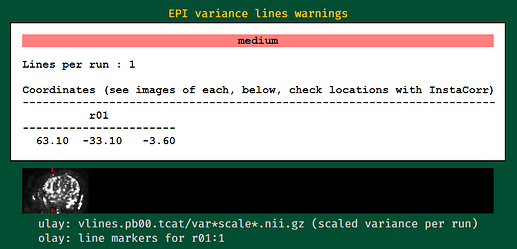
EPI variance lines warning
Sincerely,
Yang
Hi, Yang-
OK, thanks for explaining those points. I guess we had covered the cost function item in a different thread.
The EPI-anatomical alignment does look pretty reasonable to me, actually, as much as I can tell. It is more challenging both to align and to verify because of the low contrast, but I think that is doing a reasonable job. In the VMPFC part of the brain (frontal+inferior region, above the eyes and sinuses), I think there is probably signal dropout, and that is why the EPI looks effectively missing there. That is a common thing in FMRI, that most people will observe when looking at their data in detail like this. If you need data in that region for your study, you would have to do additional work to get an acquisition to get more data there (and likely in the subcortical regions).
I wouldn't worry about the left-right flip things here---I don't think there is enough signal to differentiate by cost function, nor is nmi the best cost function to use for it. But the visual comparison looks better for standard EPI-anatomical alignment.
The variance line warnings can happen as false positives at times, which is why we include the visual check. Edges of high variance can trigger a warning, but indeed I don't see any here.
--pt
1 Like
Hi ptaylor,
I replaced @SSwarper with sswarper2 and added the mask from freesurfer, which outputs some errors, but the result seems to be fine. Is this may be related to SSW align history record? BTW, It's really great to record SSW align history!!!
SSW align hist: Allin_mask_ss
++ Prepare for running @djunct_edgy_align_check (ver = 1.51)
++ Copy refbox (/home/ylz/abin/MNI152_2009_template_SSW.nii.gz[1]) to workdir
++ 3dcalc: AFNI version=AFNI_24.0.19 (Mar 29 2024) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset /media/ylz/B0EA93B7EA9377F4/postgraduate/fMRIanalysis/MI.learning/data/bids/sub-04/anat/ssw_align_hist.sub-04/__workdir_EAC_Y1F6zeSK4kx/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_24.0.19 (Mar 29 2024) [64-bit]
++ Authored by: A cast of thousands
** FATAL ERROR: can't open dataset /media/ylz/B0EA93B7EA9377F4/postgraduate/fMRIanalysis/MI.learning/data/bids/sub-04/anat/junk_sswAllin_mask_ss.nii
** Program compile date = Mar 29 2024
** FATAL ERROR: Can't open dataset 'eac_0_cp.nii'
** Program compile date = Mar 29 2024
++ 3dAllineate: AFNI version=AFNI_24.0.19 (Mar 29 2024) [64-bit]
++ Authored by: Zhark the Registrator
** FATAL ERROR: can't open -master dataset '/media/ylz/B0EA93B7EA9377F4/postgraduate/fMRIanalysis/MI.learning/data/bids/sub-04/anat/ssw_align_hist.sub-04/__workdir_EAC_Y1F6zeSK4kx/eac_1_ulay_shrp.nii' :-(
** Program compile date = Mar 29 2024
++ 3dAutomask: AFNI version=AFNI_24.0.19 (Mar 29 2024) [64-bit]
++ Authored by: Emperor Zhark
** FATAL ERROR: Can't open dataset 'eac_2_res.nii'
** Program compile date = Mar 29 2024
++ 3dMedianFilter: AFNI version=AFNI_24.0.19 (Mar 29 2024) [64-bit]
** FATAL ERROR: Can't open dataset 'eac_2_res.nii'
** Program compile date = Mar 29 2024
++ 3dedge3: AFNI version=AFNI_24.0.19 (Mar 29 2024) [64-bit]
** ERROR: can't open dataset (null)
** FATAL ERROR: Can't open dataset 'eac_5_edgy.nii'
** Program compile date = Mar 29 2024
vvv: Subscript out of range.
2dcat: No match.
+* Removing temporary files '/media/ylz/B0EA93B7EA9377F4/postgraduate/fMRIanalysis/MI.learning/data/bids/sub-04/anat/ssw_align_hist.sub-04/__ssw_intermed_qdOnsOUSVn2*'
rm: No match.
Thanks,
Yang
Hi, Yang-
Hmm. I wonder if there is an earlier error from running the program? Do you save hte full text output from the terminal when running sswarper2, such as doing one of the following to make a "log_ssw2.txt" text file of all terminal text:
# tcsh syntax
sswarper2 .... |& tee log_ssw2.txt
# bash syntax
sswarper2 ... 2>&1 | tee log_ssw2.txt
?
One thing to note: when you have a mask to add in, FreeSurfer outputs its results to a 256x256x256 grid, and that will likely not match that of your input data. You will want to resample the mask to match the grid of your input data. If you have no obliquity in either the data you input to FS's recon-all or the input to AFNI's sswarper2, then that can be done just by:
3dresample -input MASK_FS -master DSET_ANAT -prefix MASK_FS_RES
NB also, you should like use the mask that @SUMA_Make_Spec_FS makes using FS output, which tends to be similar but cleaner than recon-all's.
FreeSurfer applies obliquity when it processed data, as well. That is fine to do, but would add another wrinkle about trying to match the grid of the FS mask with an anatomical that has obliquity in it (as well as being on a different grid). That is why we strongly recommend deobliquing the anatomical dataset before running either FreeSurfer or sswarper2---it makes combining their outputs afterwards much easier.
--pt
Hi, ptaylor
Thanks for replying so quickly!
First, there is no earlier error from running the program, only have a warning below. And I saved full text output from the terminal.
+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
I used the same T1w data to FS's recon-all and AFNI's sswarper2 generated by Dimon. And I've already run @SUMA_Make_Spec_FS. The resampling was done by 3dAllineate. My script as follow:
Dimon -infile_list uniq_image_list.txt \
-gert_create_dataset \
-gert_write_as_nifti \
-gert_to3d_prefix ${subj}_T1w \
-gert_outdir $bids_dir/$subj/anat \
-dicom_org \
-use_last_elem \
-save_details Dimon.details \
-use_obl_origin \
-gert_quit_on_err
3dAllineate \
-1Dmatrix_apply IDENTITY \
-final NN \
-source ${sdir_suma}/fs_parc_wb_mask.nii* \
-master $idir/${subj}_T1w.nii \
-prefix $odir/mask_${subj}.nii.gz
time sswarper2 \
-base ${SSW_template} \
-subid ${subj} \
-mask_ss $odir/mask_${subj}.nii.gz \
-input $idir/${subj}_T1w.nii \
-odir $odir
I don't see oblique infomaton output by 3dinfo (maybe here Data Axes Tilt: Plumb)
3dinfo sub-01_T1w.nii
++ 3dinfo: AFNI version=AFNI_24.0.19 (Mar 29 2024) [64-bit]
Dataset File: /media/ylz/B0EA93B7EA9377F4/postgraduate/fMRIanalysis/MI.learning/data/bids/sub-01/anat/sub-01_T1w.nii
Identifier Code: AFN_LGSuqovakcIcJTiTPqvWWQ Creation Date: Wed Apr 3 20:26:37 2024
Template Space: ORIG
Dataset Type: Anat Bucket (-abuc)
Byte Order: LSB_FIRST {assumed} [this CPU native = LSB_FIRST]
Storage Mode: NIFTI
Storage Space: 23,068,672 (23 million) bytes
Geometry String: "MATRIX(0,0,-1,87.5688,1,0,0,-152.28,0,-1,0,119.4004):256,256,176"
Data Axes Tilt: Plumb
Data Axes Orientation:
first (x) = Anterior-to-Posterior
second (y) = Superior-to-Inferior
third (z) = Left-to-Right [-orient ASL]
R-to-L extent: -87.431 [R] -to- 87.569 [L] -step- 1.000 mm [176 voxels]
A-to-P extent: -152.280 [A] -to- 102.720 [P] -step- 1.000 mm [256 voxels]
I-to-S extent: -135.600 [I] -to- 119.400 [S] -step- 1.000 mm [256 voxels]
Number of values stored at each pixel = 1
-- At sub-brick #0 '?' datum type is short: 0 to 4181
----- HISTORY -----
[ylz@yoyimiya: Wed Apr 3 20:26:37 2024] to3d -quit_on_err -prefix sub-01_T1w.nii -use_last_elem IM0 IM1 IM2 ... IM174 IM175
Thanks,
Yang
Hm, OK. that looks reasonable, indeed.
And yes, seeing Plumb and looking at the matrix above it, it looks like there is not obliquity present. You can also run:
3dinfo -is_oblique DSET
to verify.
To check that the grids match (which the program should check, anyways), you can run:
3dinfo -same_all_grid DSET1 DSET2
If you see all 1s output, you are good (each represents a property being checked, as described in the help file).
I'm running an example locally now to see if I get the same behavior, will ping back.
--pt
Thanks for providing 3dinfo with these helpful tips!
I check the oblique on T1w data, the grids match between T1w and fs_parc_wb_mask, the grids match between T1w and resampled mask, that all looks good on the last one (-same obl).
3dinfo -is_oblique sub-01_T1w.nii
0
3dinfo -same_all_grid sub-01_T1w.nii ../../derivatives/sswarper2/sub-01/mask_sub-01.nii.gz
1 1 1 1 1
1 1 1 1 1
3dinfo -same_all_grid sub-01_T1w.nii ../../derivatives/recon-all/sub-01/SUMA/fs_parc_wb_mask.nii.gz
0 1 0 0 1
0 1 0 0 1
--Yang
Hi, Yang-
Great, that looks good, indeed.
After an initial look into this error message, there is good news/bad news. First, the bad news: there is a filename mistake in the script. The good news is that this just affects the image making for an intermediate QC image, not the actual processing itself.
So, thanks for bringing this up, and I will work on adding a fix in shortly. But you should be fine continuing your processing.
--pt
1 Like
Hi, ptaylor
Glad to see what the problem is, it didn't seem like a big problem. Thank you very much for your answers!
Yang