AFNI version info (afni -ver): AFNI_24.0.01
Dear AFNI,
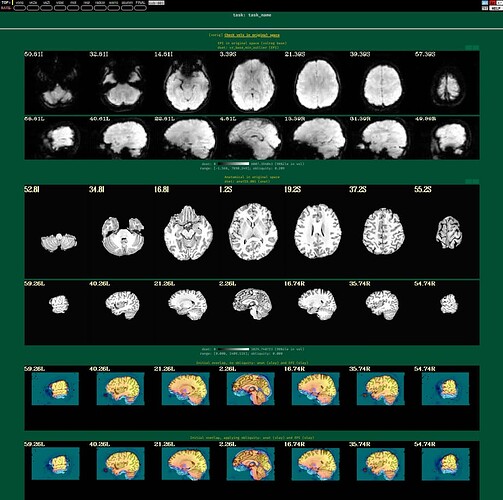
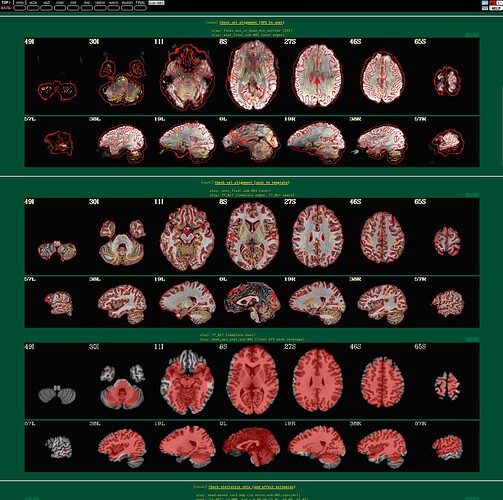
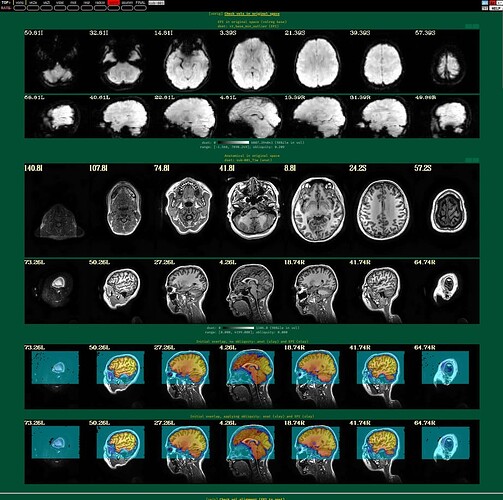
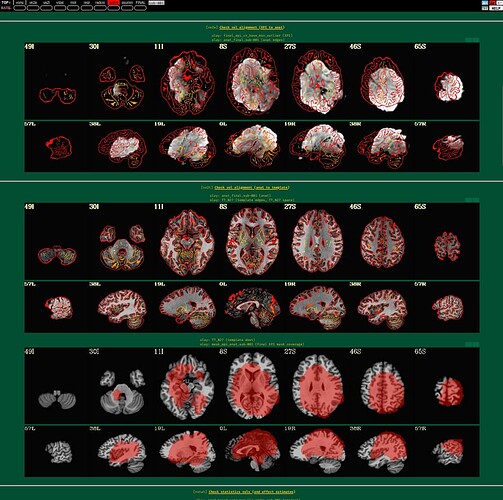
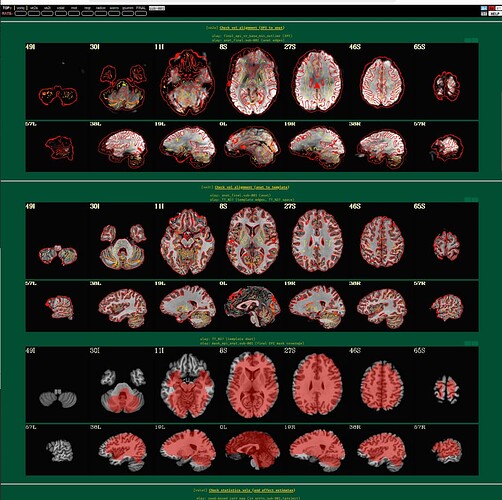
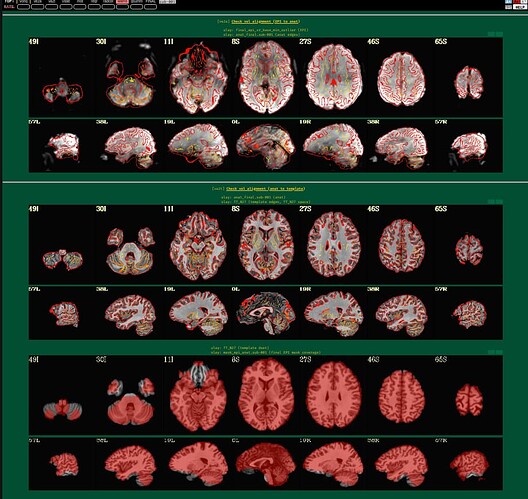
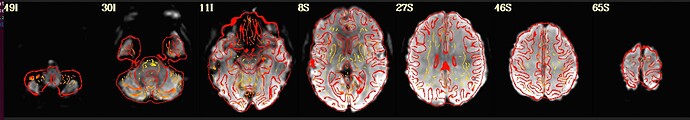
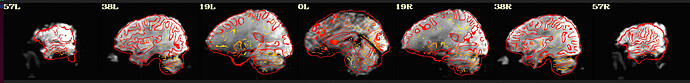
I'm using afni to preprocess my resting state data. I tried to run it twice, the first time following example 11, the second time with some deletions, but the QC report shows a poor alignment of epi to anat.
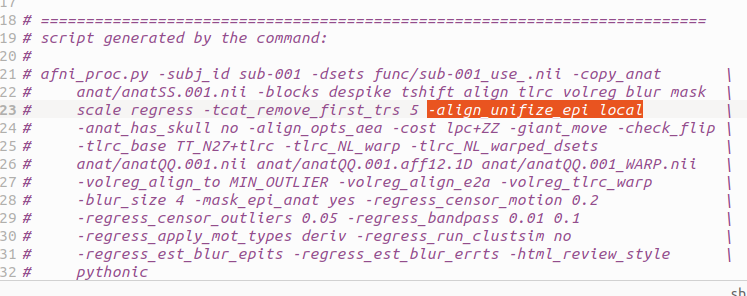
The first scipt:
afni_proc.py \
-subj_id sub-${subj} \
-dsets func/sub-${subj}_use_.nii \
-copy_anat anat/anatSS.${subj}.nii \
-blocks despike tshift align tlrc volreg blur \
mask scale regress \
-tcat_remove_first_trs 5 \
-anat_has_skull no \
-align_opts_aea -cost lpc+ZZ \
-giant_move \
-check_flip \
-tlrc_base $template \
-tlrc_NL_warp \
-tlrc_NL_warped_dsets anat/anatQQ.${subj}.nii \
anat/anatQQ.${subj}.aff12.1D \
anat/anatQQ.${subj}_WARP.nii \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-blur_size 4 \
-mask_epi_anat yes \
-regress_censor_motion 0.2 \
-regress_censor_outliers 0.05 \
-regress_bandpass 0.01 0.1 \
-regress_apply_mot_types deriv \
-regress_run_clustsim no \
-regress_est_blur_epits \
-regress_est_blur_errts \
-html_review_style pythonic
The second script:
afni_proc.py \
-subj_id sub-001 \
-blocks despike tshift align tlrc volreg blur \
mask scale regress \
-dsets func/sub-001_use_.nii \
-copy_anat anat/sub-001_T1w.nii \
-anat_has_skull yes \
-tcat_remove_first_trs 5 \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-align_opts_aea -cost lpc+ZZ \
-giant_move -check_flip \
-tlrc_base TT_N27+tlrc \
-tlrc_NL_warp \
-mask_epi_anat yes \
-regress_motion_per_run \
-blur_size 4 \
-regress_bandpass 0.01 0.1 \
-regress_censor_motion 0.2 \
-regress_censor_outliers 0.05 \
-regress_apply_mot_types deriv \
-regress_est_blur_detrend yes \
-regress_run_clustsim no \
-html_review_style pythonic \
-execute
The anat to template alignment is fine, but ve2a are showing a bad alignment.
This is the first time I've used afni_proc for preprocessing, is there an error in script?
I am very appreciate that if there have any reply!
Thanks,
Yang