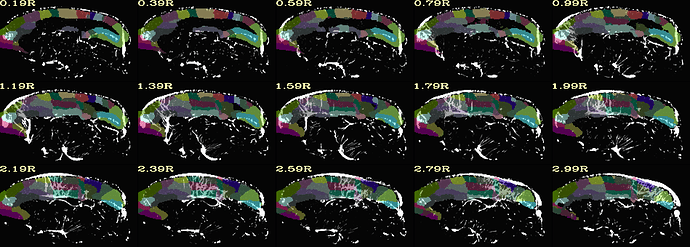
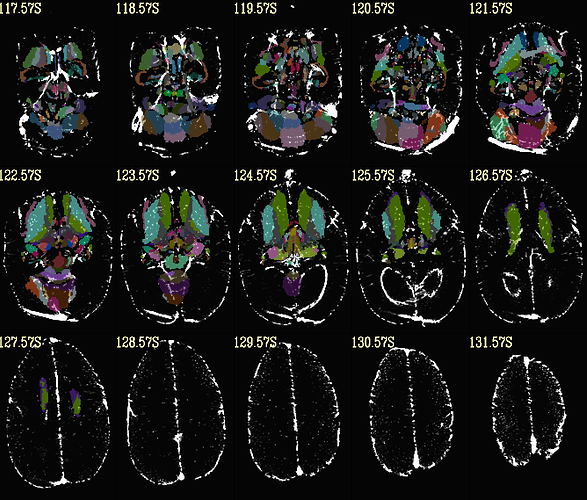
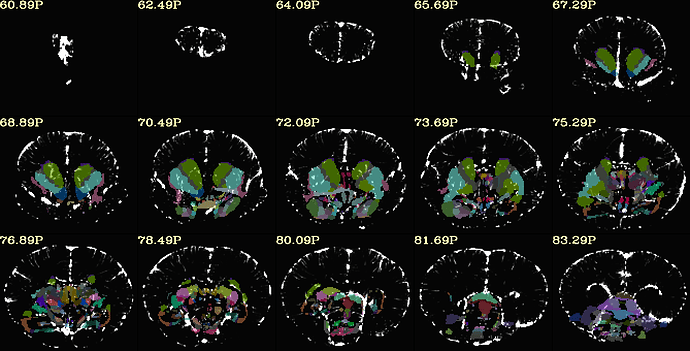
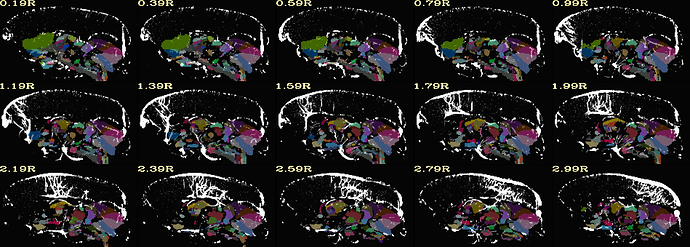
Hi, it's me again!
I managed to align CT brain (vascular signal only) to MRI template (MBM). Then, I wanted to use SAM (the subcortical atlas of marmoset), so I found that SAM has been integrated into MBM through SAM_T2_template.nii.gz. Then I tried to align CT brain to SAM_T2_template.nii.gz, but I was not successful.
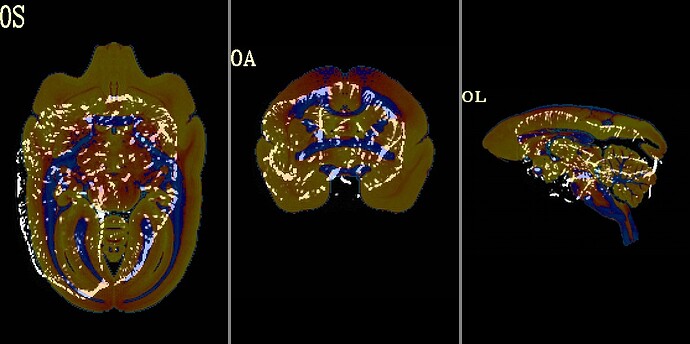
here is init_qc_03.input_NL+base when aligning with SAM_T2_template.nii.gz
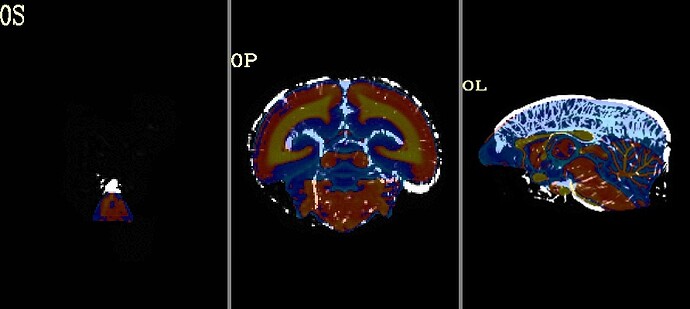
here is the same QC when aligning with MBM t2 template. (much better right?)
I guess the extra brain stem part in SAM_T2_template.nii.gz confuses the program. But maybe this problem can be solved by adding a brain mask?
The most important question I want to ask is, currently SAM_T2_template.nii.gz is 0.15x0.15x-0.15, and it has different origins than MBM templates/atlas. I want to run animal_warper once with both the cortical (MBM) and subcortical (SAM) atlases, but now they have different voxel size, shape, and origin, what's the best way to do it? I tried to resample SAM_T2_template.nii.gz/MBM template/atlas, not good.
here is the information on SAM
Template Space: SAM_Marmoset
Dataset Type: Anat Bucket (-abuc)
Byte Order: LSB_FIRST {assumed} [this CPU native = LSB_FIRST]
Storage Mode: NIFTI
Storage Space: 20,870,016 (21 million) bytes
Geometry String: "MATRIX(-0.15,0,0,13.65,0,-0.15,0,27,0,0,0.15,-18.9):184,278,204"
Data Axes Tilt: Plumb
Data Axes Orientation:
first (x) = Left-to-Right
second (y) = Posterior-to-Anterior
third (z) = Inferior-to-Superior [-orient LPI]
R-to-L extent: -13.800 [R] -to- 13.650 [L] -step- 0.150 mm [184 voxels]
A-to-P extent: -14.550 [A] -to- 27.000 [P] -step- 0.150 mm [278 voxels]
I-to-S extent: -18.900 [I] -to- 11.550 [S] -step- 0.150 mm [204 voxels]
Number of values stored at each pixel = 1
-- At sub-brick #0 '?' datum type is short: 0 to 976
here is the information on MBM template
Template Space: ORIG
Dataset Type: Anat Bucket (-abuc)
Byte Order: LSB_FIRST {assumed} [this CPU native = LSB_FIRST]
Storage Mode: NIFTI
Storage Space: 15,876,000 (16 million) bytes
Geometry String: "MATRIX(-0.2,0,0,14.6,0,-0.2,0,14.4,0,0,0.2,-3.6):147,200,135"
Data Axes Tilt: Plumb
Data Axes Orientation:
first (x) = Left-to-Right
second (y) = Posterior-to-Anterior
third (z) = Inferior-to-Superior [-orient LPI]
R-to-L extent: -14.600 [R] -to- 14.600 [L] -step- 0.200 mm [147 voxels]
A-to-P extent: -25.400 [A] -to- 14.400 [P] -step- 0.200 mm [200 voxels]
I-to-S extent: -3.600 [I] -to- 23.200 [S] -step- 0.200 mm [135 voxels]
Number of values stored at each pixel = 1
-- At sub-brick #0 '?' datum type is float: 0 to 4.12392
Thank you!
Yibei