Hi, Peng-
Thanks, I ran the following script for your just dcm2niix_afni-converted dataset:
#!/bin/tcsh
# Potential script to change a dataset acquired in sphinx orientation (e.g.,
# like many animal scan acquisitions) to human-like orientation.
#
# NB: we have to check whether RSP or LSP is more appropriate, to know
# that left/right ends up being correct
#
# This script does not overwrite files, so please move or remove tmp*
# files before running, if they exist already
#
# ---------------------------------------------------------------------------
# input dataset, presumably just after raw file conversion
set dset_in = dicom_T2_TurboRARE_20230607141101_20001.nii
# What is the appropriate orientation for refitting, from RAI orient?
# NB: for most sphinx-to-human acquisitions, I would typically expect
# to use either RSP or LSP for the final orientation here.
set new_ori = RIP # could also be LIP: check with external information
# ---------------------------------------------------------------------------
# First, check for presence of obliquity
set is_obl = `3dinfo -is_oblique "${dset_in}"`
if ( ${is_obl} ) then
echo "+* WARNING: input dataset ${dset_in} is oblique."
echo " Obliquity will *not* be preserved here"
else
echo "++ Processing (non-oblique) dataset: ${dset_in}"
endif
# check for pre-existing run
if ( -f tmp_00_RAI.nii.gz || -f tmp_01_${new_ori}.nii.gz ) then
echo '** ERROR: pre-existing tmp_0*.nii.gz files. Will not overwrite.'
echo ' Please move or remove them before running.'
exit 1
endif
# Things are easiest starting from a standard RAI orientation---this
# step updates both data and header information, so no "corrective" action
# occurs here (but any obliquity will not be preserved here)
3dresample \
-orient RAI \
-input "${dset_in}" \
-prefix tmp_00_RAI.nii.gz
# Copy the data before refitting, because refitting changes the header
# in place
3dcopy \
tmp_00_RAI.nii.gz \
tmp_01_${new_ori}.nii.gz
# ... and update the header information, effectively reorienting the
# dataset
3drefit \
-orient ${new_ori} \
tmp_01_${new_ori}.nii.gz
echo "++ DONE. Please visually verify results for final dataset:"
echo " tmp_01_${new_ori}.nii.gz"
echo " Please take extra caution to verify left/right correctness."
The output looked reasonable for reorientating. NB: In most sphinx-to-humanlike reorienting, I am used to refitting RAI to RSP or to LSP; in this case, I had to convert it to RIP or LIP, because the brain looked upside down otherwise. This might be due to the particular scan setup, but I just wanted to note it for future uses of this script. Also note that that script removes obliquity information from the header as it goes; if that is important to preserve, we could discuss how to do so, but it is more complicated.
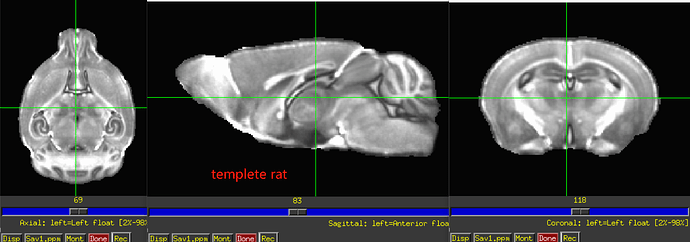
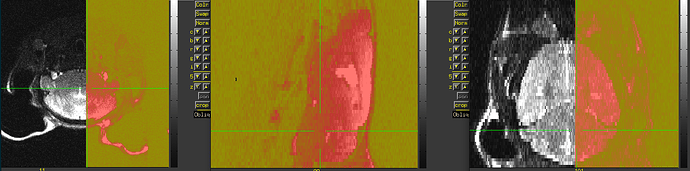
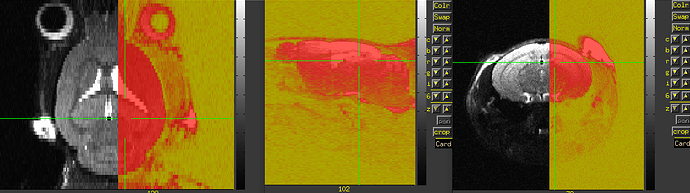
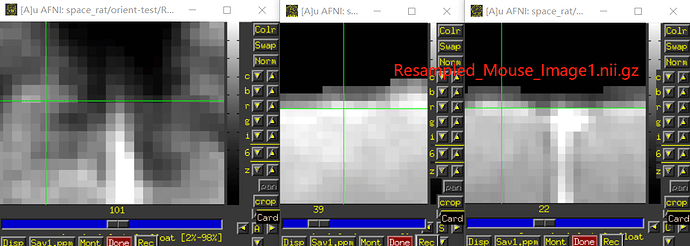
Also, I think you will have to still verify left/right correctness somehow. For example, do you know how the animal was placed in the scanner to start with, so you can tell what is the animal's physiological left side in the original acquisition? For example, here is an image of your original data as underlay, with the three panels in axial (left=left), sagittal (left=anterior) and coronal (left=left) orientation, and the overlay is a copy of the dataset with have of the FOV zeroed out:
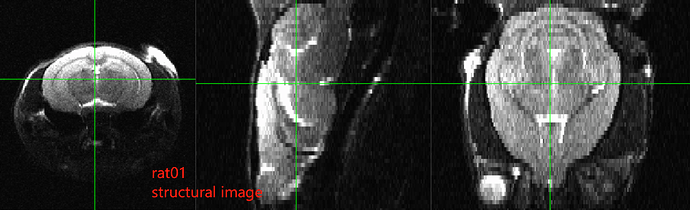
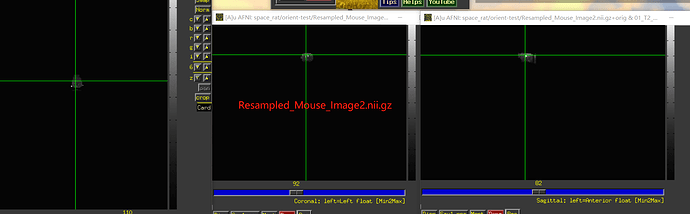
After running the script as written above on each of those datasets, the results look like:
Now, the question is, in the original dataset, can you tell whether the zeroed-out half is the anatomical left or right? If the original zeroed out half is the anatomical left, then things appear to be correct currently; otherwise, you would want to change the value of
new_ori in the script to
LIP.
--pt