AFNI version info (afni -ver):
AFNI_21.1.07 'Domitian'
First I want to thank you all for creating this software and being so active on this forum. I really appreciate it.
Here's my issue: when I run the below script 3dMSS_test.sh (inspired by example 5 from the 3dMSS documentation:
3dMSS -prefix output -jobs 16 \
-lme 'pain+stimulus+s(TENT,k=9)+s(TENT,k=9,by=pain)' \
-ranEff 'list(subject=~1)' \
-qVars 'pain,stimulus,TENT' \
-prediction @HRF.table \
-dataTable @extended_df_final.txt
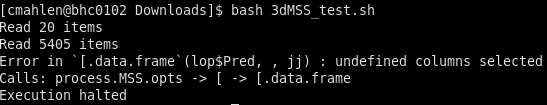
I get the following error:
Read 20 items
Read 7567 items
Error in if (len%%wd != 0) errex.AFNI(paste("The content under -dataTable is not rectangular !", :
argument is of length zero"
Calls: process.MSS.opts
Execution halted
I tried following the advice from this thread (appending \ to each line and creating different subject ids for different conditions), and this thread (using file_tool -test -infile … to check for bad formatting → returns "script has 0 bad characters")
Here is a snippet from extended_df_final.txt (the actual .txt file contains a consistent use of \t, whereas here it is formatted differently for readability):
path subject pain stimulus TENT
/path/to/brick[0] sub-1432 chronicbackpain taste 0
/path/to/brick[1] sub-1432 chronicbackpain taste 1
/path/to/brick[2] sub-1432 chronicbackpain taste 2
...
/path/to/brick[9] sub-1432 chronicbackpain tasteless 0
/path/to/brick[10] sub-1432 chronicbackpain tasteless 1
/path/to/brick[11] sub-1432 chronicbackpain tasteless 2
...
/path/to/brick[0] sub-1234 healthy taste 0
/path/to/brick[1] sub-1234 healthy taste 1
/path/to/brick[2] sub-1234 healthy taste 2
...
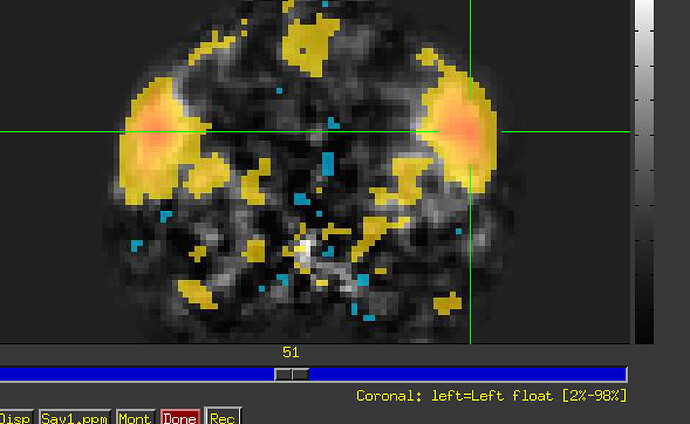
Repeating with TR 0-8 for each subject and each condition taste and tasteless. brick is the bucket output of 3dDeconvolve which contains beta maps for each TENT function for both taste and tasteless conditions (I can provide that script as well if needed). The output from 3dDeconvolve makes sense (or so I think)--after using 3dbucket to aggregate subbricks, I'm able to look at the HRF for both taste and tasteless conditions for each voxel in the brain.
I'm not sure what to do to resolve the "rectangular" error. I've written code to ensure that all of the rows are filled without success (the file seems rectangular).
Any help would be greatly appreciated!
I should also clarify my design: subjects either have chronicbackpain or are healthy. In the MRI, they receive a stimulus that has taste or a control tasteless stimulus. Our TR is 1.