AFNI version info (afni -ver): 23.3.14
Dear AFNI members:
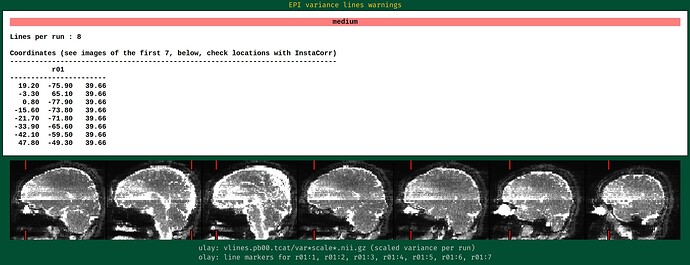
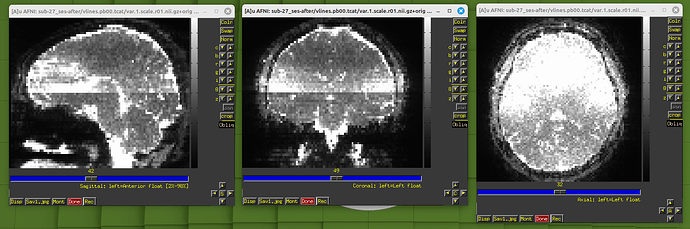
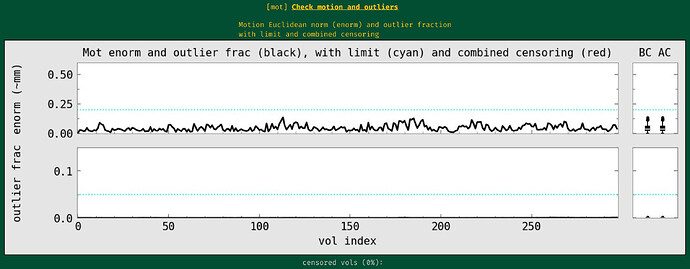
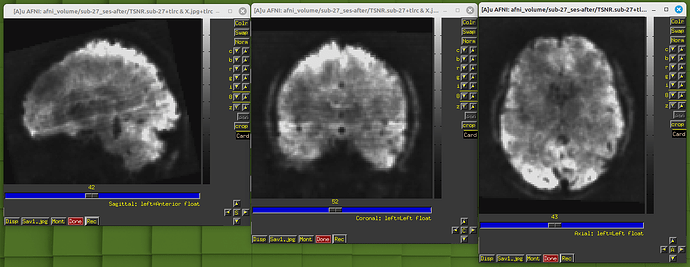
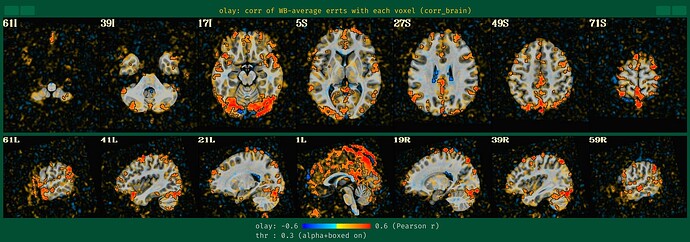
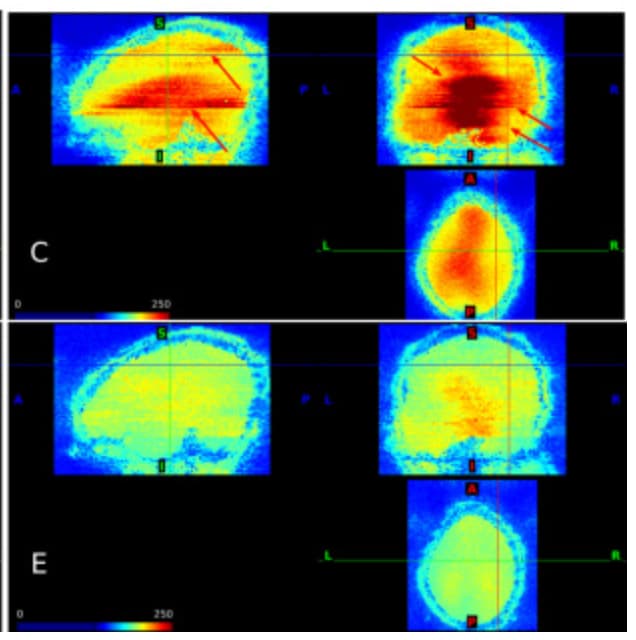
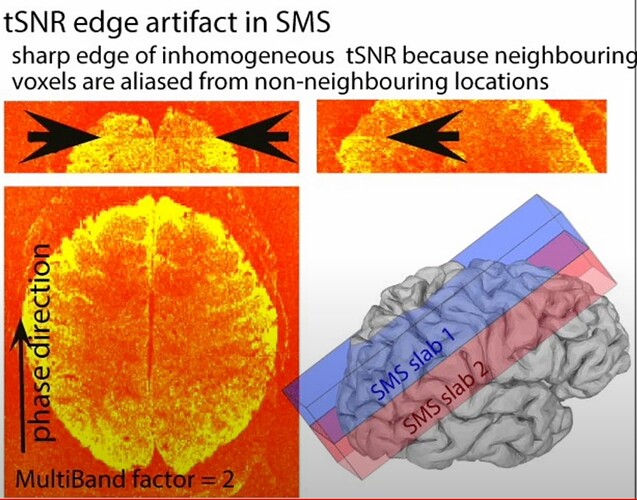
During the QC, I found my dataset shows obvious horizental variance lines though it indicates the vertical lines which are less obvious. I don't know how to explain it. I wonder whether this phenomenon correlates with some scan options, like phase encoding direction and whether this dataset is valuable to the following analysis.
PhaseEncodingDirection: J