AFNI version info (afni -ver): 23.3.14
Dear AFNI members:
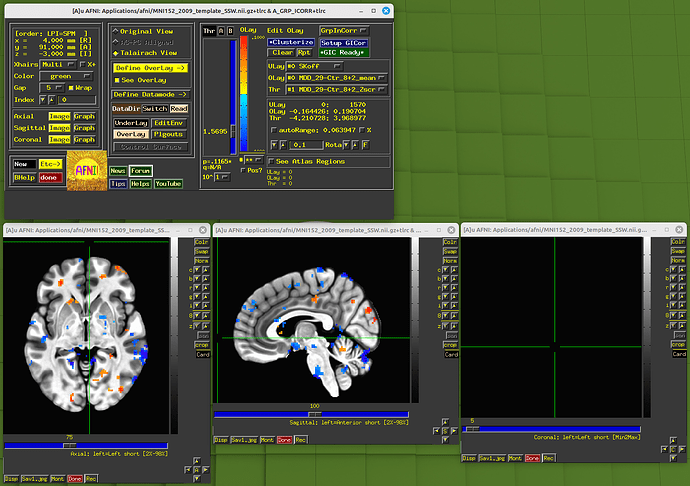
In the final QC dir, I could not find vstat.DMN and vstat.vis images. After checking the processing log, I find the following warnings:
gen_epi_review.py -script @epi_review.sub-01 -dsets pb00.sub-01.r01.tcat+orig.HEAD
cat
cat out.ap_uvars.txt
afni_python_wrapper.py -eval data_file_to_json()
gen_ss_review_scripts.py -exit0 -init_uvars_json out.ap_uvars.json -write_uvars_json out.ss_review_uvars.json
** failed to guess at any stats dset, resting state?
(else X-matrix file "X.xmat.1D" may not apply)
++ writing ss review basic: @ss_review_basic
** no stats_dset dset, cannot drive view_stats, skipping...
++ writing ss review driver: @ss_review_driver
This is my afni_proc.py script:
afni_proc.py \
-subj_id $subID \
-script proc_${subID}_${runID} \
-out_dir /home/renyiyuan/fMRI/Main/NIFTI/derivatives/afni_volume/${subID}_${runID} \
-html_review_style pythonic \
-remove_preproc_files \
-execute \
\
-dsets /home/renyiyuan/fMRI/Main/NIFTI/$subID/$runID/func/${subID}_${runID}_task-rest_bold.nii.gz \
-copy_anat /home/renyiyuan/fMRI/Main/NIFTI/derivatives/freesurfer/$subID/SUMA/brainmask.nii.gz \
-anat_has_skull no \
-anat_follower anat_w_skull anat /home/renyiyuan/fMRI/Main/NIFTI/derivatives/freesurfer/$subID/SUMA/T1.nii.gz \
-anat_follower_ROI aaseg anat /home/renyiyuan/fMRI/Main/NIFTI/derivatives/freesurfer/$subID/SUMA/aparc.a2009s+aseg_REN_all.nii.gz \
-anat_follower_ROI aeseg epi /home/renyiyuan/fMRI/Main/NIFTI/derivatives/freesurfer/$subID/SUMA/aparc.a2009s+aseg_REN_all.nii.gz \
-anat_follower_ROI FSVe epi /home/renyiyuan/fMRI/Main/NIFTI/derivatives/freesurfer/$subID/SUMA/fs_ap_latvent.nii.gz \
-anat_follower_ROI FSWe epi /home/renyiyuan/fMRI/Main/NIFTI/derivatives/freesurfer/$subID/SUMA/fs_ap_wm.nii.gz \
-anat_follower_erode FSVe FSWe \
\
-blocks despike tshift align tlrc volreg blur mask scale regress \
-radial_correlate_blocks tcat volreg regress \
\
-tcat_remove_first_trs 2 \
\
-tshift_align_to -tzero 1 \
\
-align_opts_aea -cost lpc+ZZ -giant_move -check_flip \
-align_unifize_epi local \
\
-tlrc_base MNI152_2009_template_SSW.nii.gz \
-tlrc_NL_warp \
\
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
\
-blur_size 4 \
\
-mask_epi_anat yes \
\
-regress_motion_per_run \
-regress_apply_mot_types demean deriv \
-regress_censor_motion 0.2 \
-regress_censor_outliers 0.05 \
-regress_anaticor_fast \
-regress_anaticor_label FSWe \
-regress_ROI_PC FSVe 3 \
-regress_ROI_PC_per_run FSVe \
-regress_make_corr_vols aeseg FSVe FSWe \
-regress_est_blur_epits \
-regress_est_blur_errts \
Because of low download speed, I manually installed the AFNI packages and completed the afni_system_check.py. The AFNI_ATLAS_PATH env arg has been set in .afnirc.
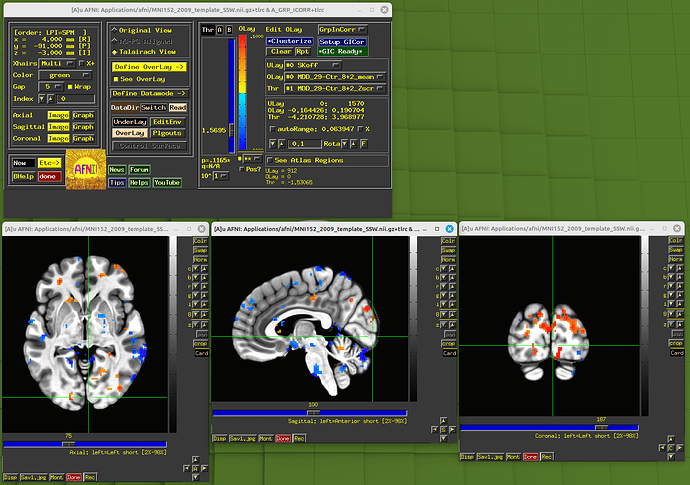
By the way, there is another relevant question. In the paper, the vstat.vis is based on the seed (4R, 91A, 3I in MNI space). But in my practice, that seems a wrong position. See the picture:
If I change the MNI x, y, z from 4, 91, -3 to 4, -91, -3, the position is correct.
Is there anything I misunderstand?
Howdy-
Re. the location of the seed for APQC HTML:
- by default, AFNI uses RAI-DICOM coordinate system. This goes back to the 90s, using DICOM's coordinate convention. RAI means that being rightward, anterior or inferior to the coordinate origin has a negative coordinate.
- The default coordinate system order in the GUI is RAI, and this is shown in the upper left corner of the GUI.
- On your system, you have changed this to be the LPI-SPM system. Therefore, in your GUI setup, you will see leftward, posterior and inferior coordinates having negative values, relative to the coordinate origin.
- In the paper you are citing, the coordinates were left in default RAI-DICOM convention.
See here in the GUI guide of the documentation about that part of the GUI.
In MNI-2009c space, the seed-based locations in RAI-DICOM notation are:
# x y z space roi_label netw
# -----------------------------------------------------------------------------
5 49 40 MNI lh-PCC DMN
-4 91 -3 MNI rh-cort-vis Vis
64 12 2 MNI lh-cort-aud Aud
To translate these to your chosen LPI-SPM coordinate setup, you would flip the sign on the given x- and y-coords when jumping to them in the GUI, or change your GUI setup.
Re. APQC HTML not having a stats dset:
That surprises me, and I might ask to see your full output* text log of the processing to help see if there were any intermediate problems. One thing I see is that you are using the -remove_preproc_files option, and this will do the following:
-remove_preproc_files : delete pre-processed data
At the end of the output script, delete the intermediate data (to
save disk space). Delete dfile*, outcount*, pb* and rm*.
This removes some important intermediate files for QC generation, actually. There won't be a stats file, anyways, but some of the things like the TSNR of the volreg data, and perhaps some other things. So, if you remove this option, you should get the full QC output.
--pt
... and actually, I have seen that there is a deeper problem with the vstat seedbased correlation map in the current version of AFNI. When using the MNI152_2009 space as a reference template space, the seedbased maps won't be generated, indeed. The space name was updated in the file to be more specific, but not in the code to match.
That has been updated/fixed in the code now, and I am starting a build, so that in a couple hours it should be available. Updating AFNI versions to 24.0.03 (and higher) should be fine for this behavior.
Sorry for the hassle, and thanks very much for bringing this up.
--pt
Hi @ptaylor,
I am glade to see this important improvement. Thank you!
For visual network seed (4R, 91A, 3I), if we enter it in a RAI-DICOM system, the x, y, z should be -4, -91, -3, right? But that is a wrong position. The position your post here is correct. So I think this is a typo in the article.
@ptaylor,
I want to know there are any potential problems with the results produced by this old version software. You know I don't want to redo the whole preprocessing. It took me a lot of time.
Howdy-
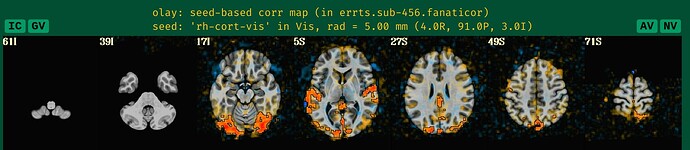
Wow, you are right about the coordinates and what was stated in the text of that article. That is somewhat blush-worthy on our part, because the text above the APQC HTML image for that figure quite clearly states it correctly:
Indeed, we should have written: (4R, 91P, 3I), and
- in RAI coord system, this would be: (-4, 91, -3)
- in LPI coord system, this would be: (4, -91, -3).
There should not be a problem with the processing results here. The issue is only with APQC HTML figure generation. You can actually regenerate the APQC HTML for a subject without re-running afni_proc.py again. If you were in the results directory output by afni_proc.py with subject ID ${subj}, you would run:
apqc_make_tcsh.py -subj_dir . -uvar_json out.ss_review_uvars.json
apqc_make_html.py -qc_dir QC_${subj}
That will make a new QC directory with the name provided after -qc_dir .., and if there was a pre-existing one (which there will be in this case), that pre-existing one will get moved first to a backup name: QC_${subj}_TIMESTAMP, where TIMESTAMP is the time stamp of when you started generating the new one, something like: 2023-08-22-17-13-40.
So, when you have updated code, you can do this. to only regenerate the APQC HTML for that subject.
--pt
OK, thing is going to be alright. Thanks for your kind help again.
Totally, and thanks for mentioning these points. They are useful for us to know and fix.
--pt