Hi Paul and Daniel,
thanks again for your instructions. I took some time to play around with your options. For the IsoSurface, I think coloring the convexity is the most straight forward way for me and worked really well! However, when I assigned the colors of my liking to the ROIs, I noticed that this decision did not get saved when using the "Save View" option from the Viewer in order to reproduce what I did before (changing dset to convexity, picking a colormap and a corresponding color and so on). Is this the intended behavior?
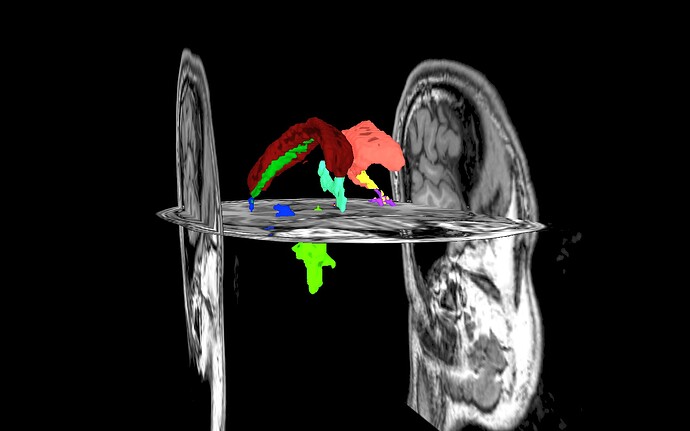
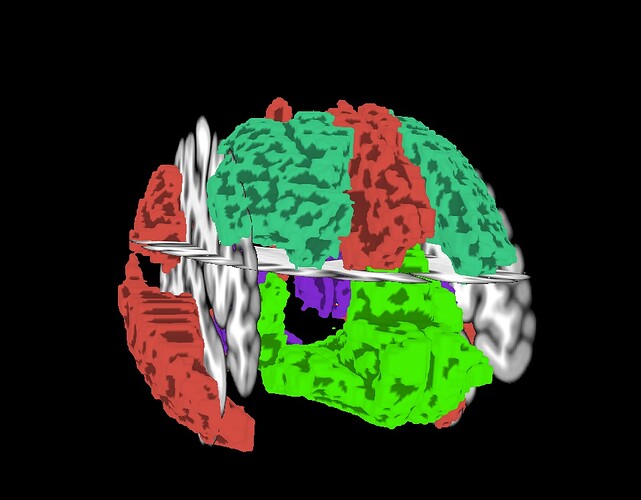
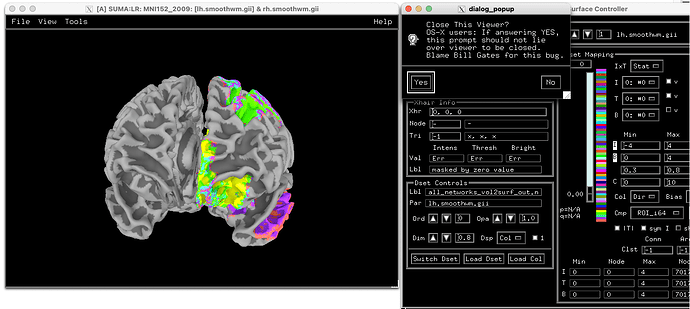
For displaying the volumetric ROIs on the surface, I was not as lucky.
I tried 3dVol2Surf using the preprocessed MNI surface kindly provided by AFNI like this:
3dVol2Surf -spec ../data/MNI152_2009_surf/MNI152_2009_both.spec \
-surf_A ../data/MNI152_2009_surf/lh.pial.gii -surf_B ../data/MNI152_2009_surf/lh.smoothwm.gii \
-sv ../data/MNI152_2009_surf/MNI152_2009_SurfVol.nii -grid_parent ../../charles_BNA_mask_as_networks_1mm+tlrc \
-map_func ave -f_steps 50 -f_index nodes \
-out_niml all_networks_vol2surf_out.niml.dset
Apart from the fact that only the left hemisphere is now colored (I guess this is intended behavior running 3dVol2Surf like that but preferrably, I would like to see it on both hemispheres), even selecting a classical colormap for this purpose like ROI_i64, I get different colors.
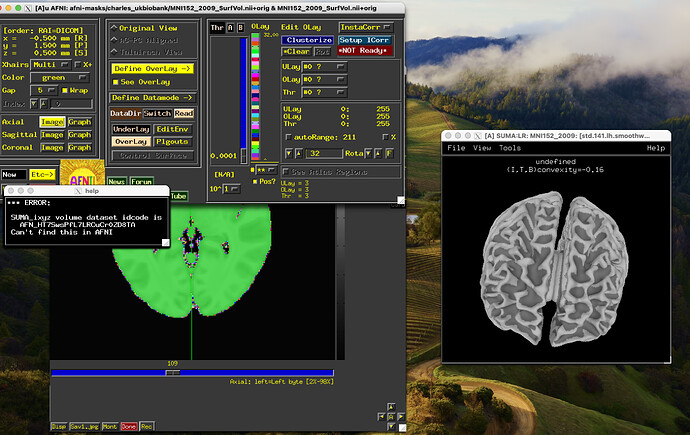
And unfortunately, with the straight forward option that I also though would get me the result - running both AFNI and SUMA with -niml option and having them talk to each other - nothing really happened. But I do get some error messages that the the idcode of the suma dataset was not found in AFNI.
I launched AFNI and SUMA with
afni -niml &
suma -niml -spec ../data/MNI152_2009_surf/MNI152_2009_both.spec -sv ../data/MNI152_2009_surf/MNI152_2009_SurfVol.nii &
I guess this somehow has to do with my mask not having any relationship with the preprocessed MNI surface data provided by AFNI?
And one final issue (as if this wasn't enough already): If I try to my fun rotating brain recording as a .mpeg file, the console tells me a successful output, but the file is nowhere to be found, which is quite odd. I have noticed that AFNI uses the version of ffmpeg that I had installed on my mac before to compile the video, which is different from what I saw in the bootcamp video. Maybe it has something to do with that?
++ Running '/opt/homebrew/bin/ffmpeg -loglevel quiet -y -r 24 -f image2 -i realmovie.EXH.%06d.ppm -b 400k -qscale 11 -intra realmovie.mpg' to produce realmovie.mpg
. **DONE**
As always, I have learned a lot in the process and understood some fundamentals, especially with the AFNI academy video you kindly referenced, Paul.
Warm regards,
Jonas



![[AFNI Academy] Advanced Visualization - atlases as surfaces in SUMA](https://discuss.afni.nimh.nih.gov/uploads/default/original/2X/6/691fc9d41c8d5d5fb1dcec6facd85574e2a1bdeb.jpeg)