AFNI version info (afni -ver): Version AFNI_24.1.01 'Publius Septimius Geta'
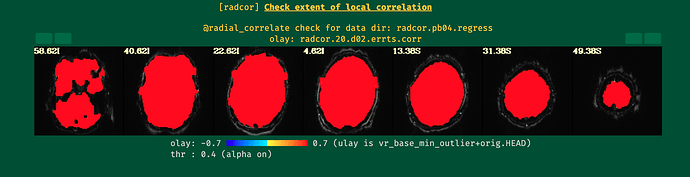
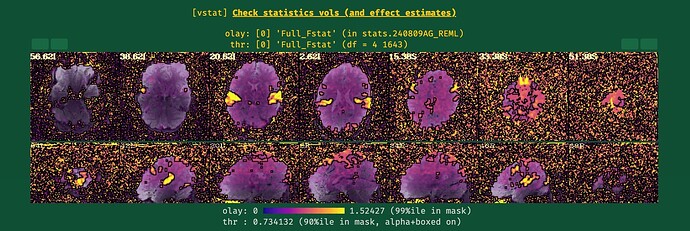
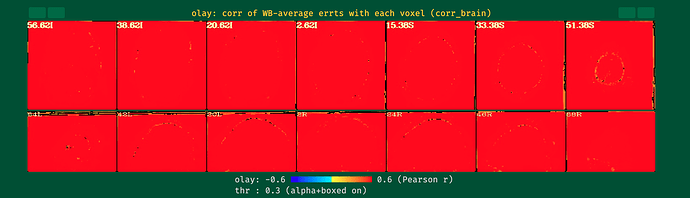
I ran the first round of processing of my data without blurring and motion correction (i.e., using only blocks: volreg, mask, scale, regress), and my data looked pretty normal. However, after I included blur and censoring (and related flags) as in the script below, some of the QC metrics went completely wild (see the attached screenshots below). I'm a newbie when it comes to AFNI, so perhaps I'm missing something obvious here.
Do you have any ideas about what might be causing this and why?
I've tried the following so far and here's what I found:
- Same script (blur and censor), but with the automask flag removed; it yields the same error.
- Same script, but with censoring removed; blur works fine with automask. TSNR improves, but the local correlation increases significantly compared to the previous processing without blur.
- Same script, but with censor outliers added; I get the same unusual results.
- Same script, but only with blur (no automask or censoring); I still see strange artifacts.
It seems that the combination of options / flags I use is causing this, but I can't tell what. Any help with this would be much appreciated, thanks!
afni_proc.py -subj_id ${subj} \
-script afni_proc_${subj}_prosody_task.tcsh \
-out_dir afni_proc_${subj}_prosody_task.results \
-dsets ${subj}_MB_prosody_run1.nii ${subj}_MB_prosody_run2.nii \
${subj}_MB_prosody_run3.nii ${subj}_MB_prosody_run4.nii \
-blocks volreg blur mask scale regress \
-tcat_remove_first_trs 6 \
-blur_filter -1blur_fwhm \
-blur_size 4 \
-blur_in_automask \
-volreg_align_to MIN_OUTLIER \
-regress_reml_exec \
-regress_local_times \
-regress_censor_motion 0.3 \
-regress_opts_3dD \
-num_stimts 4 \
-stim_label 1 consistent75 \
-stim_times_AM1 1 $tdir/240809AG_consistent75_exact.txt 'dmUBLOCK(1)' \
-stim_label 2 consistent100 \
-stim_times_AM1 2 $tdir/240809AG_consistent100_exact.txt 'dmUBLOCK(1)' \
-stim_label 3 conflicting75 \
-stim_times_AM1 3 $tdir/240809AG_conflicting75_exact.txt 'dmUBLOCK(1)' \
-stim_label 4 conflicting100 \
-stim_times_AM1 4 $tdir/240809AG_conflicting100_exact.txt 'dmUBLOCK(1)' \
-gltsym 'SYM: +consistent75 + consistent100 -conflicting75 -conflicting100' \
-glt_label 1 'ConsistentConflicting' \
-tout \
-execute