AFNI version info (afni -ver):
align_epi_anat.py \
-anat2epi -anat sub-756055_run-01_T1+orig \
-save_skullstrip -suffix _al_junk \
-epi vr_base_min_outlier+orig -epi_base 0 \
-cost lpc+ZZ -giant_move -cmass cmass -check_flip \
-align_centers yes -volreg off -tshift off
I ran afni_proc.py with the below option.
-align_opts_aea -cost lpc+ZZ -giant_move -cmass cmass -check_flip
I found the alignment between epi and T1w image worse. So, I checked the script generated from afni_proc.py and tested align_epi_anat.py. I concluded that the larger difference in cmass between the two images made this issue. For the reason, I tried below four.
- run align_epi_anat.py with the option "-align_centers yes".
--> Still gave me an error about the large cmass difference.
- run align_epi_anat.py with the option "-align_centers on"
--> Still gave me an error about the large cmass difference.
- run align_epi_anat.py with the option "ginormous_move"
--> Still gave me an error about the large cmass difference.
- run @Align_Centers first between epi and T1w images and run align_epi_anat.py with the T1w image and center-changed epi image without the option "align_centers".
--> it worked very well, but this seems to be too difficult to automatize the preprocess because afni_proc.py does not have the option to run @Align_Centers before align_epi_anat.py.
So, my questions are
- What's the difference between -align_centers yes and on.
- What's the purpose of the option align_centers? I wonder this because the consideration for the too large cmass differences seems to be earlier than the application of align_centers.
- Do you have any good idea for this huge cmass difference issue?
Just you know, I am working on HCP data.
Thanks!
Howdy-
Thanks for the details. To have a bit of backstory, could you share some of the APQC HTML images related to this?
It would help to see the following images from under the "vorig" QC block (I will show example images here from the AFNI Bootcamp demo data, for subject "FT"):
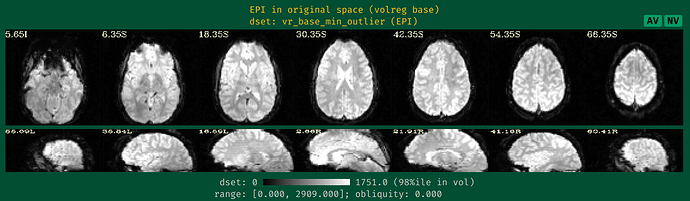
- the EPI montage (so we can see the coverage and tissue contrast):
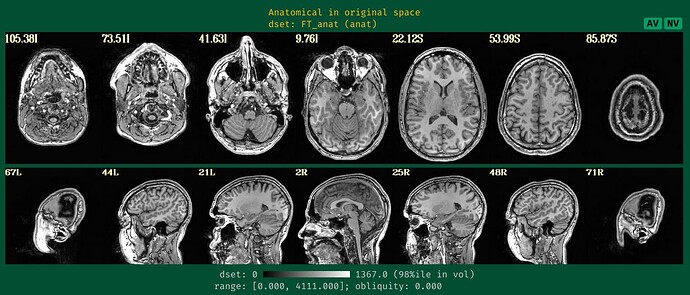
- the anatomical montage (so we can see the coverage and tissue contrast):
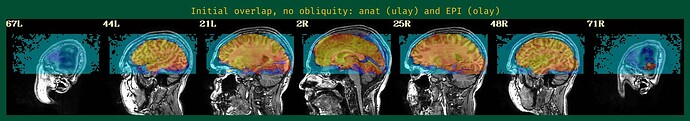
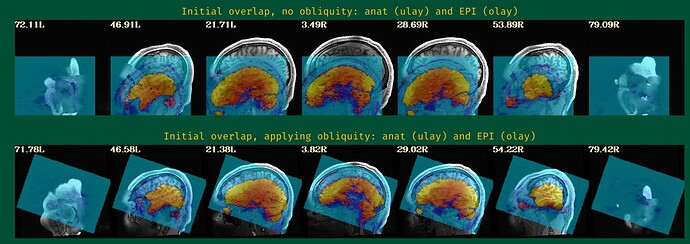
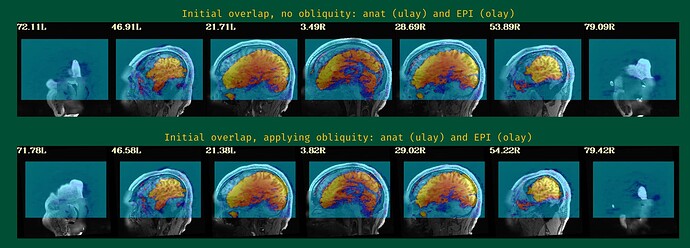
- The EPI-anatomical initial overlap (might have 2 images, if obliquity is present---please include both):
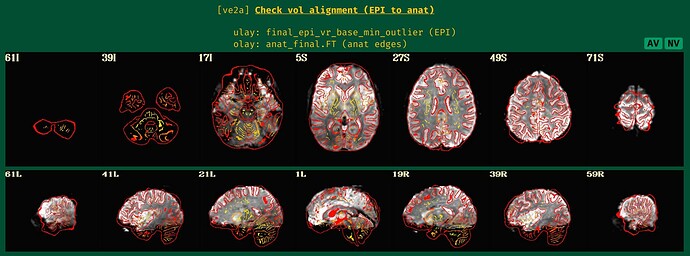
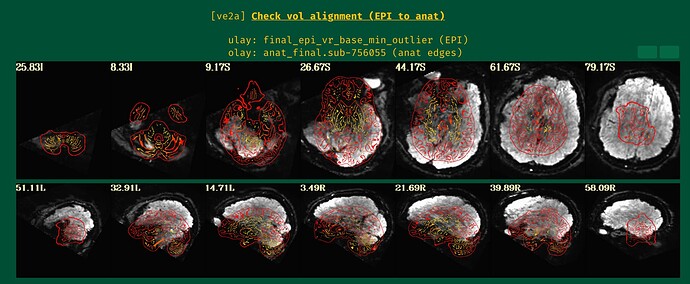
... and one more image from the "ve2a" block:
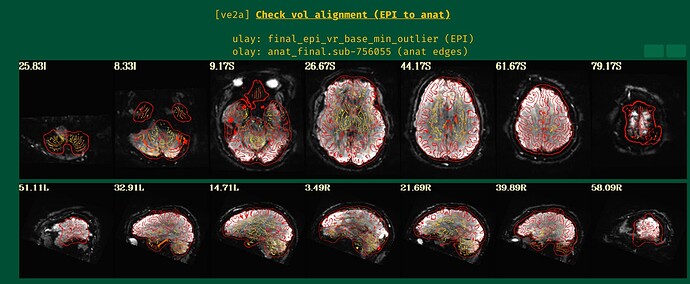
- The result of the alignment, with anatomical edges shown over the target EPI:
Also, since this is for human data, does your afni_proc.py command include this option:
-align_unifize_epi local
? We recommend including that for all human data now typically, since that helps overcome EPI brightness inhomogenity that is so often present (and even if not present, doesn't seem to hurt).
thanks,
pt
1 Like
Thanks. I tested with the option. It indeed enhanced the alignment quality. But, it still could not solve the error about the larger cmass difference.
This was my afni_proc.py options.
afni_proc.py -subj_id $subj \
-script $script_dir/proc.$subj -scr_overwrite \
-out_dir $out_dir \
-dsets \
$epi_dir/${subj}_task-${task}_acq-${acq}_run-01_bold.nii.gz \
$epi_dir/${subj}_task-${task}_acq-${acq}_run-02_bold.nii.gz \
-copy_anat $anat_dir/${subj}_run-01_T1w.nii.gz \
-anat_has_skull yes \
-anat_follower anat_w_skull anat $anat_dir/${subj}_run-01_T1w.nii.gz \
-tcat_remove_first_trs $ign \
-blocks tshift align volreg mask surf blur scale regress \
-align_opts_aea -cost lpc+ZZ -giant_move -cmass cmass -check_flip -align_centers yes \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-mask_epi_anat yes \
-mask_segment_anat yes \
-mask_segment_erode yes \
-surf_anat ${fs_dir}/${subj}/surf/SUMA/${subj}_SurfVol.nii \
-surf_spec ${fs_dir}/${subj}/surf/SUMA/std.60.${subj}_lh.spec \
${fs_dir}/${subj}/surf/SUMA/std.60.${subj}_rh.spec \
-blur_size $fwhm \
-regress_censor_motion $motion_thr \
-regress_censor_outliers 0.05 \
-regress_bandpass 0.01 0.1 \
-regress_ROI WMe CSFe \
-regress_apply_mot_types demean deriv \
-regress_motion_per_run \
-regress_opts_3dD -jobs $n_jobs_per_subj \
-html_review_style pythonic \
> $script_dir/log/log.$subj.txt
Output
I ran afni_proc.py with the same option after applying @Align_Centers to epi scan based on T1w images.
Output
With the option -align_unifize_epi local,
<U: final_epi_vr_base_min_outlier, O:anat_final.sub-756055>
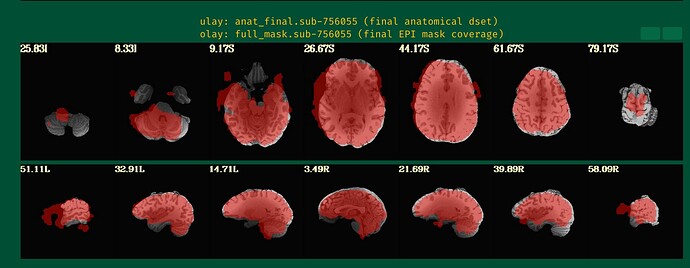
<U: anat_final.sub-756055 O:full_mask.sub-756055>
Thanks for your reply again,
Jin.
Hi, Jin-
Thanks for sharing both the current code and the images, that helps.
And for a bit of background on alignment considerations, we have this set of talks, if that's of interest.
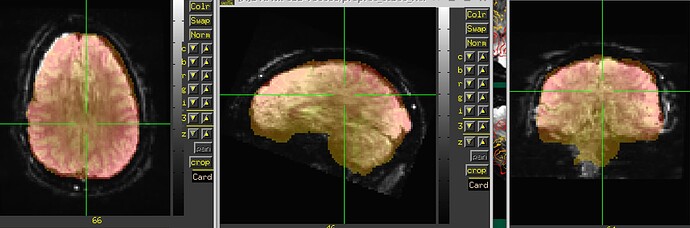
The starting point of the EPI-anatomical alignment is going to start from the "Initial overlap" images at the bottom of the "vorig" section. When there is no obliquity, there is just the direct overlap; when obliquity is present, it will be the lower of the two images (when obliquity is applied). I don't see a raw anatomical image, so I don't know if that dataset has obliquity or whether it is just the EPI (the usual case, when present), but I will assume that only the EPI has obliquity.
- To check, you can run
3dinfo -obliquity DSET and see whether the answer is nonzero; the obliquity value is also reported beneath each of the EPI and anatomical dataset images in the "vorig" block of the QC HTML here. Since you are using FreeSurfer output for surfaces, it would be important to know if the anatomical here does have any obliquity; FreeSurfer and AFNI treat obliquity differently, so it would be better to remove it before any processing.
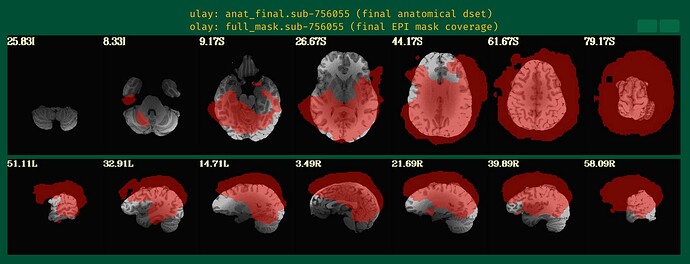
From the images showing the mask over the anatomical, I can see that the anatomical has a bit of brightness gradient. Normally, we would use sswarper2 to both skullstrip the anatomical and to calculate the nonlinear alignment to the template space; the program also unifizes the brightness of the anatomical when skullstripping. Even though you are not sending your data to a template space, that might help a bit. Your -copy_anat .. dset would be the one without skull (and then you would use -anat_has_skull no, rather then "yes"), and the -anat_follower anat_w_skull anat .. would be the one with skull, to be available for later visualization checks.
Since your EPI and anatomical have pretty good initial overlap (again, focused on the lower of hte "Initial overlap" images), I don't think you should need -align_centers yes or recentering the -cmass ... I would try this just this for align block opts:
-align_unifize_epi local \
-align_opts_aea -cost lpc+ZZ -big_move -check_flip \
The -big_move opens up the parameter space of searching a bit (allowing bigger shift), but I don't think a very large shift is necessary.
--pt
1 Like