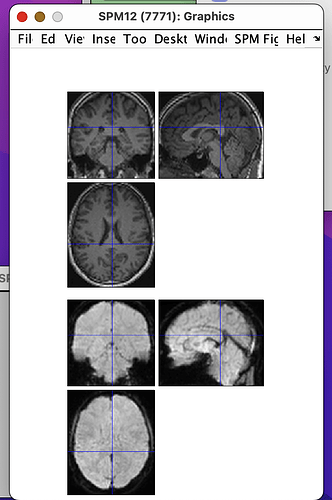
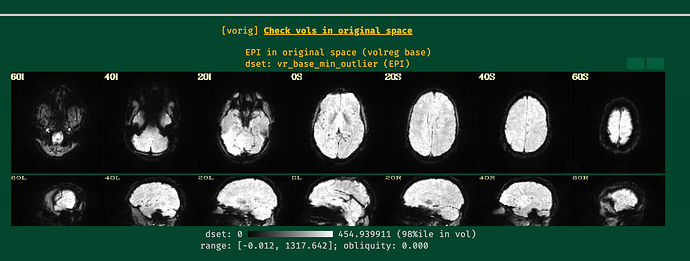
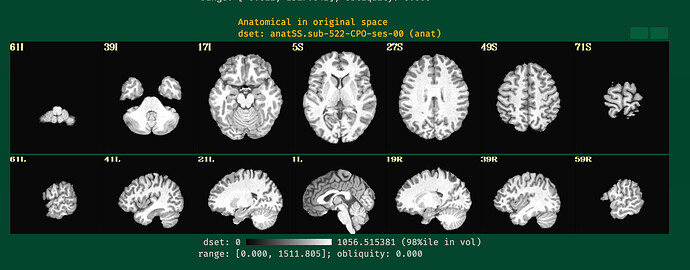
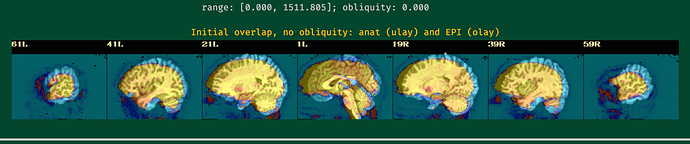
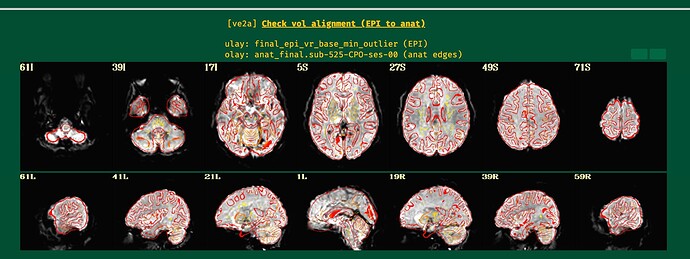
Hi all,
I just wanted to update this thread for future viewers. It turned out the root of my problems was the cost function in this line:
-align_opts_aea -cost lpc+ZZ -big_move -align_centers yes -cmass cmass.
After some playing around with by the AFNI gurus, the correction I went with was:
-align_opts_aea -cost nmi -giant_move -check_flip.
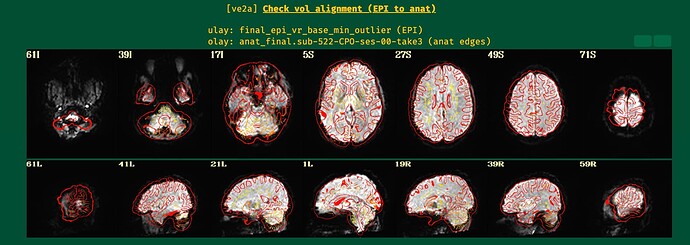
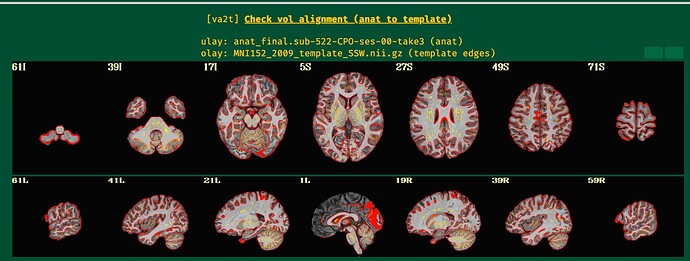
While lpc+ZZ is still the first cost function to try, when the EPI has lower tissue contrast then nmi might be a good fallback to try if the EPI-anatomical alignment does not go well, as here.
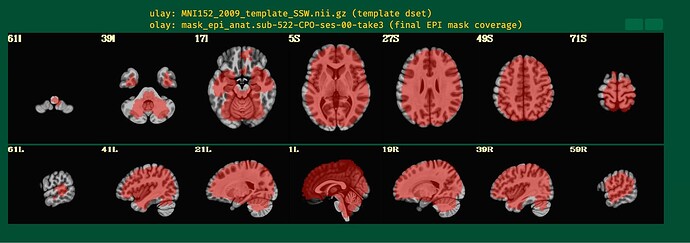
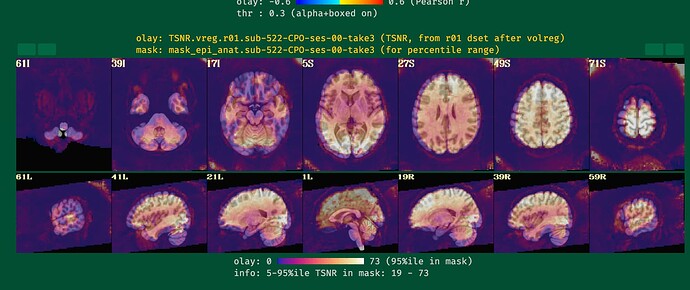
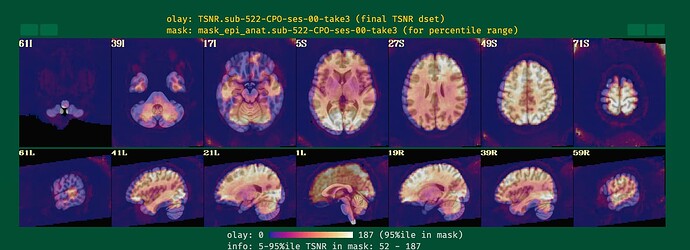
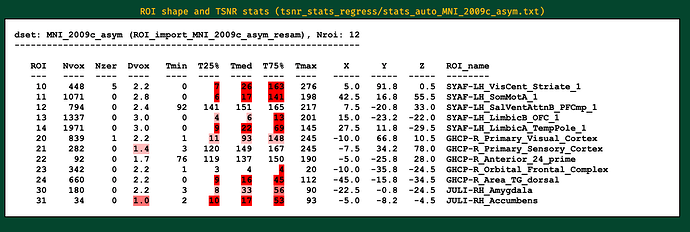
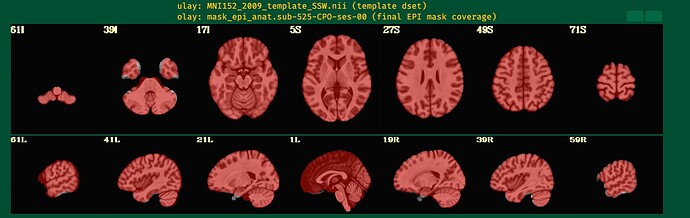
I will post the resulting picture below.
Separate comment on Example 11:
At the bottom, I will put my fully functioning script based on Example 11.
I also wanted to add to this thread, as I struggled to answer some questions about example 11 for Resting-State Preprocessing, and I think it could be helpful for future learners if they are spelled out here. I was struggling to get my epi and anat data lined up, in part due to misunderstanding these lines: (bolded were my areas of confusion)
-anat_follower **anat_w_skull anat** $SUBJECT_DIR/sub-525-CPO/ses-07/anatU.sub-525-CPO-ses-07.nii \
-anat_follower_ROI **aaseg anat** $SUBJECT_DIR/sub-525-CPO/ses-07/recon_all/sub-525-CPO-ses-07/SUMA/aparc.a2009s+aseg_REN_all.nii \
-anat_follower_ROI **aeseg epi** $SUBJECT_DIR/sub-525-CPO/ses-07/recon_all/sub-525-CPO-ses-07/SUMA/aparc.a2009s+aseg_REN_all.nii \
-anat_follower_ROI FSvent **epi** $SUBJECT_DIR/sub-525-CPO/ses-07/recon_all/sub-525-CPO-ses-07/SUMA/fs_ap_latvent.nii \
-anat_follower_ROI FSWe **epi** $SUBJECT_DIR/sub-525-CPO/ses-07/recon_all/sub-525-CPO-ses-07/SUMA/fs_ap_wm.nii \
Below is my correspondence with AFNI:
bekah:
One question I have is does this:
-anat_follower anat_w_skull anat anatU.FT.nii \
mean I need to put in three files after the -anat_follower (the anat with skull aka the unprocessed T1, then the anatSS file, and then the anatU file), or are anat_w_skull and anat modifiers for the file anatU?
afni:
No need for anything else here. After anat_follower comes 3 things, like you have: label grid_type dset_name. The anat_w_skull is the descriptive label, and anat specifies the grid type, and the anatU.FT.nii is the dataset to follow along.
bekah:
Second, on this line:
-anat_follower_ROI aeseg epi \ aparc.a2009s+aseg_REN_all.nii.gz \
I am confused if I am supposed to map the epi (RS data) with native space parcellations of the epi data? Or does this mean that -anat_follower_ROI needs the aeseg file from the anatomical FreeSurfer/recon_all parcellation, as well as the epi (RS data), and then this aparc.a2009s+aseg_REN_all.nii.gz file?
afni:
This looks fine to me. The aeseg will be the label (what we use in examples, this is fine), and epi means that the follower dataset will end up on the final EPI's grid, which is likely what you want for a parcellation to apply to the EPI data.
Full working script based on Example 11, the only difference is in the align cost function and I made my motion censoring threshold more liberal:
#!/bin/bash
export SUBJECT_DIR="/Volumes/EHD/CPO_DATA"
echo "SUBJECT_DIR is set to: $SUBJECT_DIR"
/Users/bekah/abin/afni_proc.py \
-subj_id sub-525-CPO-ses-07 \
-out_dir $SUBJECT_DIR/sub-525-CPO/ses-07/preprocessed/ \
-blocks despike tshift align tlrc volreg blur mask scale regress \
-radial_correlate_blocks tcat volreg regress \
-dsets $SUBJECT_DIR/sub-525-CPO/ses-07/sub_525_CPO_ses_07_RS_-_FMRI.nii \
-blip_forward_dset $SUBJECT_DIR/sub-525-CPO/ses-07/sub_525_CPO_ses_07_RS_-_FMRI.nii'[0]' \
-blip_reverse_dset $SUBJECT_DIR/sub-525-CPO/ses-07/sub_525_CPO_ses_07_P_A_RS_-_FMRI.nii'[0]' \
-copy_anat $SUBJECT_DIR/sub-525-CPO/ses-07/anatSS.sub-525-CPO-ses-07.nii \
-anat_has_skull no \
-anat_follower anat_w_skull anat $SUBJECT_DIR/sub-525-CPO/ses-07/anatU.sub-525-CPO-ses-07.nii \
-anat_follower_ROI aaseg anat $SUBJECT_DIR/sub-525-CPO/ses-07/recon_all/sub-525-CPO-ses-07/SUMA/aparc.a2009s+aseg_REN_all.nii \
-anat_follower_ROI aeseg epi $SUBJECT_DIR/sub-525-CPO/ses-07/recon_all/sub-525-CPO-ses-07/SUMA/aparc.a2009s+aseg_REN_all.nii \
-anat_follower_ROI FSvent epi $SUBJECT_DIR/sub-525-CPO/ses-07/recon_all/sub-525-CPO-ses-07/SUMA/fs_ap_latvent.nii \
-anat_follower_ROI FSWe epi $SUBJECT_DIR/sub-525-CPO/ses-07/recon_all/sub-525-CPO-ses-07/SUMA/fs_ap_wm.nii \
-anat_follower_erode FSvent FSWe \
-tcat_remove_first_trs 4 \
-align_unifize_epi local \
-align_opts_aea -cost nmi -giant_move -check_flip \
-tlrc_base /Users/bekahthurn/afni_build/afni_atlases_dist/MNI152_2009_template_SSW.nii \
-tlrc_NL_warp \
-tlrc_NL_warped_dsets \
$SUBJECT_DIR/sub-525-CPO/ses-07/anatQQ.sub-525-CPO-ses-07.nii \
$SUBJECT_DIR/sub-525-CPO/ses-07/anatQQ.sub-525-CPO-ses-07.aff12.1D \
$SUBJECT_DIR/sub-525-CPO/ses-07/anatQQ.sub-525-CPO-ses-07_WARP.nii \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-mask_epi_anat yes \
-blur_size 4 \
-regress_apply_mot_types demean deriv \
-regress_motion_per_run \
-regress_anaticor_fast \
-regress_anaticor_label FSWe \
-regress_ROI_PC FSvent 3 \
-regress_ROI_PC_per_run FSvent \
-regress_censor_motion 0.4 \
-regress_censor_outliers 0.05 \
-regress_make_corr_vols aeseg FSvent \
-regress_est_blur_epits \
-regress_est_blur_errts \
-html_review_style pythonic