Due to brief Message Board outage, this issue was addressed by separate correspondence, but I'll fill in the replies+work here.
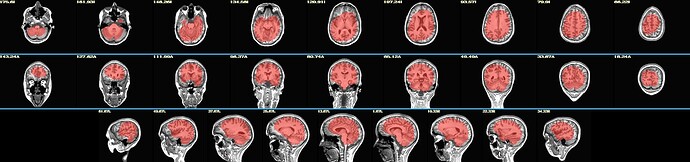
The initial sswarper2 result for the brain mask estimate was this:
... with correspondingly poor alignment to the reference template.
I initially asked to see some header info from that data, in case there is some inconsistency, specifically:
3dinfo -ad3 -obliquity -aform_real T1.nii.gz
nifti_tool -disp_hdr -infiles T1.nii.gz
The output of the nifti_tool command didn't show any weirdness, like with qform_code/sform_code values or things. The output of the 3dinfo command was:
1.953125 1.953125 2.000000 28.309 # mat44 (aform_real):
-0.027190 -0.056924 1.998956 -168.327194
1.719720 0.924484 0.052134 -199.881699
0.925485 -1.719531 -0.038145 -17.396299
... which means that there is obliquity present and the initial coordinate origin appears to be pretty offset far from the middle-ish region of the brain---therefore, initial overlap would be poor with the template, and the alignment might get lost in space. There is a directory of QC images automatically made, called ssw_align_hist*, to help identify and troubleshoot where the alignment goes wrong, which can indeed happen just at the start.
(The voxelsize is also 2x2x2mm^3, which is a bit bigger than for most anatomicals. There was another anatomical with 1x1x1mm^3 resolution that was also shared, having similar issues. In the end, we could apply the same set of steps to greatly improve results in both---see below.)
Typically, we recommend deobliquing the anatomicals before any processing (except in partial volume or other special cases), to avoid odd issues with alignment or differences in how different software deal with obliquity. Specifically, we recommend using adjunct_deob_around_origin, because it will purge obliquity while not blurring/regridding the data and also preserving the coordinate origin location. (More on the latter aspect in the present case, later.)
In the end, we developed the final script for this case. There are comments about what each step do. I had expected to need to use adjunct_deob_around_origin, as noted above. This particular case was probably a bit special that the other 3dCM and 3dAutobox were quite useful to, respectively, better centralize the coordinate origin within the brain, and to chop away many empty slices (while still keeping some padding):
#!/bin/tcsh
set subj = C1
set dset = ${subj}_T1.nii.gz
set ver = 001
set subjv = ${subj}_${ver}
set dset_pref = `3dinfo -prefix_noext "${dset}"`
# -------------------------------------------------------------------------
# deoblique, preserving coordinate origin (x,y,z) = (0,0,0)
set dset_deob = ${dset_pref}_00_DEOB.nii.gz
adjunct_deob_around_origin \
-overwrite \
-input ${dset} \
-prefix ${dset_deob}
# well, the coord origin is in an odd spot, so try recentering around CM
set dset_cm = ${dset_pref}_01_CM.nii.gz
3dcopy -overwrite ${dset_deob} ${dset_cm}
3dCM -set 0 0 0 -automask -Icent ${dset_cm}
# there is a lot of empty slices, so let's remove some for
# memory/speed considerations (but don't want it too tight)
set dset_abox = ${dset_pref}_02_ABOX.nii.gz
3dAutobox \
-overwrite \
-prefix ${dset_abox} \
-npad 10 \
-input ${dset_cm}
# can remove some lower slices, including about npad and lots of neck
set dset_zp = ${dset_pref}_03_ZP.nii.gz
3dZeropad \
-overwrite \
-prefix ${dset_zp} \
-I -30 \
${dset_abox}
# finally, let SSW work
sswarper2 \
-input ${dset_zp} \
-base MNI152_2009_template_SSW.nii.gz \
-subid ${subjv} \
-odir o.aw_${subjv} \
-cost_nl_final lpa \
|& tee log.aw_${subjv}
Full comment on the script:
I think the main issue was that the coordinates of these datasets contain obliquity, and also have hte coordinate origin (x,y,z)=(0,0,0) still reasonably far from the middle of the brain. Together, these make the initial alignments of the datasets start far off, and then some weird things can happen. These principles of alignment are discussed more in this AFNI Academy video/playlist. So, in the scripts, I first tried to remove obliquity in a way that preserves coordinate origin (using adjunct_deob_around_origin), but since that was still far from the center of mass (CM) of the brain, I just reset the coordinate origin to be at the center of mass (using 3dCM). I then noticed that there was a lot of empty space around at least one of the dsets, and that seemed like a memory waste, so I trimmed that away a bit (but still left some around, which is helpful to have) using 3dAutobox. Because there was so much sub-brain region in the inferior slices, I also used 3dZeropad to remove some of that. That doesn't have to be done, but it does save some memory when running. If you have very different datasets, the amount that could be trimmed might change.
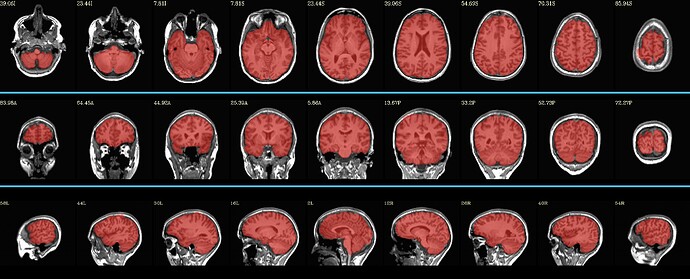
This script worked on both the dataset shown above, producing this much better brain mask:
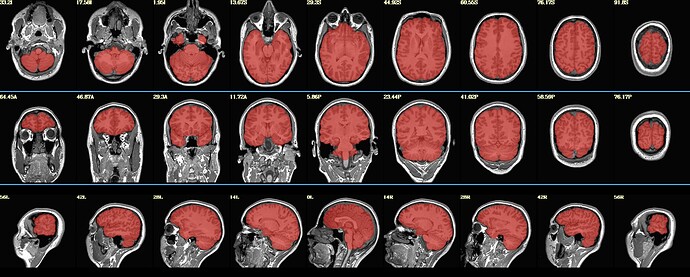
... and also on a 1x1x1mm^3 dataset, producing this brain mask:
--pt