AFNI version info (afni -ver): AFNI_23.2.11
Hi AFNI experts!
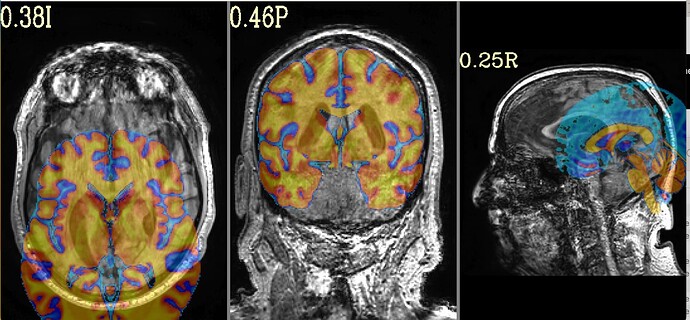
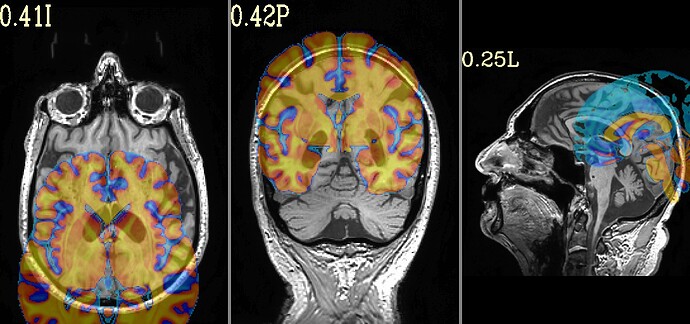
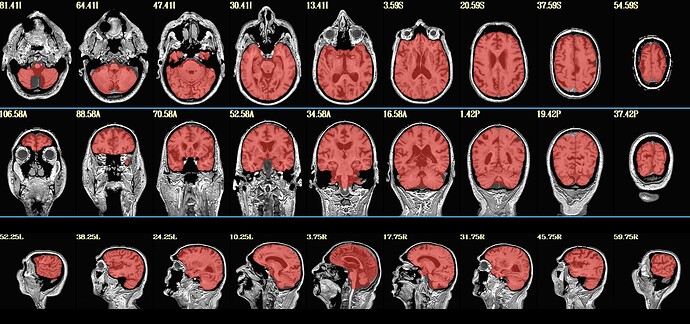
I had a question regarding interpreting SSwarper outputs. When viewing my anat outputs In afni, they seem to look good. However, when I view the "init_qc_00_overlap_usrc_obase.jpg" my anat image seems to be shifted pretty far to the right. I ran @align_centers in an attempt to get these images to align better, however as we can see from the output, the anat images are still heavily shifted. When I went and looked at my other subjects, the same issue is present.
Here is what my init_qc_00_overlap_usrc_obase.jpg looks like:
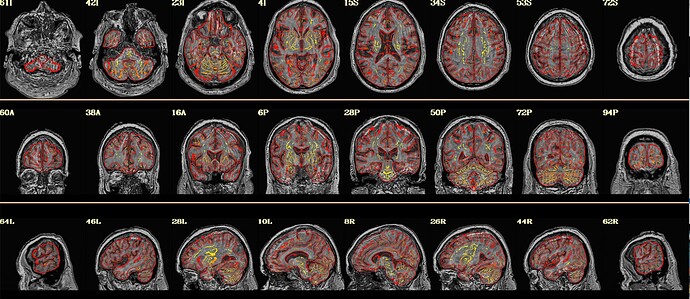
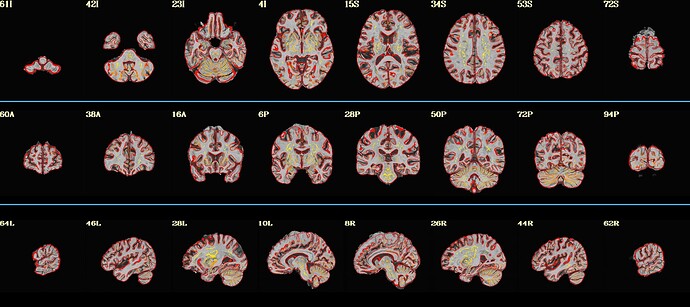
Here is what my init_qc_01_nl0.ATN005v1_ON.jpg looks like:
Here is my SSwarper script for reference:
#!/bin/bash
@SSwarper \
-input /Users/ajcoope9/Desktop/2023-PD/fMRIdata_DICOMsort/ATN005v1_ON/Nifti/"ATN005v1_ON_T1-MPRAGE-8min_20220331092729_201.nii.gz" \
-base /Users/ajcoope9/abin/MNI152_2009_template_SSW.nii.gz \
-subid 'ATN005v1_ON' \
-odir /Users/ajcoope9/Desktop/2023-PD/Afni_outputs/ATN005v1_ON/AFNI_SS_opt \
# -minp MP \
-nolite
-extra_qc_off \
-jump_to_extra_qc \
-deoblique \
-deoblique_refitly \
-warpscale 0.5 \
-SSopt 'strings' \
-aniso_off \
-ceil_off \
-echo \
-verb \
-noclean
When looking into this on google, it appears a little shifting to the right is relatively normal. However, in all the AFNI tutorials I watched online, none of their output images appeared this shifted, so I am assuming this means all of my subjects need to be redone? Any advice on this issue is greatly appreciated, thank you!