when i checked my results, i found weird results in the regress part.i dont konw how to describe it , but it looked definitely different withthe normal results, i dont know why
Hi-
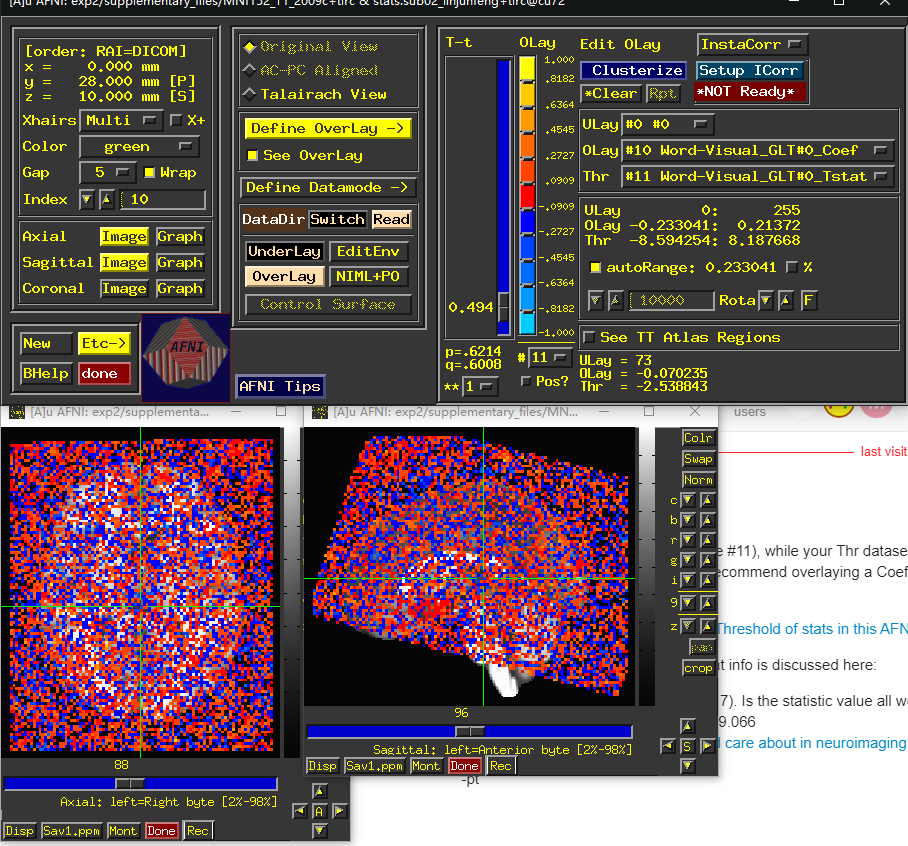
Your Olay dataset is one T-stat (volume #11), while your Thr dataset is a separate T-stat (volume #5). Those likely shouldn't be paired together like that. We would typically recommend overlaying a Coef volume (effect/point estimate) and use the corresponding T-stat volume as Thr.
Please see this discussion of Overlay/Threshold of stats in this AFNI Academy video.
This useful style of using both coef+stat info is discussed here:
- Chen G, Taylor PA, Cox RW (2017). Is the statistic value all we should care about in neuroimaging? Neuroimage. 147:952-959. doi:10.1016/j.neuroimage.2016.09.066
Is the statistic value all we should care about in neuroimaging? - PubMed
-pt
Indeed, that looks okay, except that the stats for that contrast are not great. But set the OLay coloring to also be volume 5 (or play with the threshold a bit). That will give a better feel for more of a main effect (I assume Visual is).
Note that even the Visual T-stat caps at 5 though...
- rick
thanks for your relpy very much!
but i dont think it is because i misparied the Olay and Thr image.
I change the subbrick as your recommendation, but it still remain the same pattern.....
Hm, that is odd. All the values, both the Coef and the Tstat, seem to be quite small:
- The Coef look small because the colors shown are all dark red and blue, which are in your cbar around 0.
- The Tstat look small because all throughout the brain, thresholding at |t|=0.494 (corresponding to p=0.6214) makes salt and pepper pattern of what is shown/not shown.
Did you check the Full F-stat in the stats volume (probably the [0]th volume), which you could display as both Olay and Thr (because it is pure stat and has no Coef)? That will tell you omnibus information about your fit.
What is being displayed is a GLT (general linear test), so it is derived from some directly-estimated stimuli. How do the individual stimulus results that go into (Word and Visual on their own, I am guessing?) look?
The APQC HTML created by afni_proc.py (if you processed your data with afni_proc.py) will also warn you about potential stimulus correlated motion, or other regression issues. Have you verified everything is fine, there? Some more notes on that are:
-
Taylor PA, Glen DR, Chen G, Cox RW, Hanayik T, Rorden C, Nielson DM, Rajendra JK, Reynolds RC (2024). A Set of FMRI Quality Control Tools in AFNI: Systematic, in-depth and interactive QC with afni_proc.py and more. doi: 10.1101/2024.03.27.586976.
https://www.biorxiv.org/content/10.1101/2024.03.27.586976v1 -
Reynolds RC, Taylor PA, Glen DR (2023). Quality control practices in FMRI analysis: Philosophy, methods and examples using AFNI. Front. Neurosci. 16:1073800. doi: 10.3389/fnins.2022.1073800
Frontiers | Quality control practices in FMRI analysis: Philosophy, methods and examples using AFNI
--pt
One thing to be very careful with (and which is difficult for us to help with) is the stimulus timing. If that gets messed up, the results will be garbage. For example, if there is a pre-steady state time mismatch between the timing and the EPI, or if the format you are providing does not match the usage.
If you would like, you could email the timing files and X.xmat.1D, and I could offer comments.
- rick