AFNI version info (afni -ver): 24.2.1
Hi AFNI experts!
I am preprocessing some task fMRI data that has 2 runs per session, per subject. I am aware that with afni_proc you can specify multiple runs with a wildcard * in your -dsets argument. Similarly, our stimulus timing files are in the format of the timing_tool.py, with each row/line being a separate run. If I understand correctly, the wildcard will read run-01 and run-02 in that order, and since they are in "alphabetical" order, the first row in the stimulus timing file will be applied to run-01, and the second row will be applied to run-02.
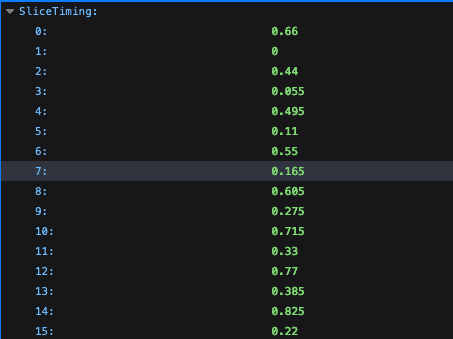
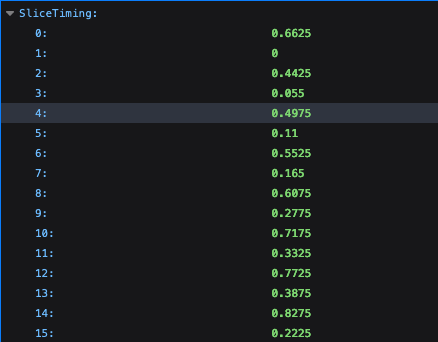
However, I am wondering what to do when you have multiple runs with regard to the slice timing files? Currently I have made separate slice timing files for each run. An example of one of my slice timing files:
0.66 0 0.44 0.055 0.495 0.11 0.55 0.165 0.605 0.275 0.715 0.33 0.77 0.385 0.825 0.22 0.66 0 0.44 0.055 0.495 0.11 0.55 0.165 0.605 0.275 0.715 0.33 0.77 0.385 0.825 0.22 0.66 0 0.44 0.055 0.495 0.11 0.55 0.165 0.605 0.275 0.715 0.33 0.77 0.385 0.825 0.22
I'm wondering, should I be setting up my slice timing file (as specified under the -tpattern option for -tshift_opts_ts) similarly to my stimulus timing files, with one row/line per run? Or is there another way that I will have to specify the correct slice timing file per run?
Below is my afni_proc.py code, if it helps.
## 3. Run afni_proc.py
apptainer exec -B /work $AFNI_path afni_proc.py \
-dsets sub-${sub_id}_ses-${ses_id}_task-${task_id}_run-*_bold.nii \
-copy_anat ${prewarp_path}/anatSS.sub-${sub_id}_ses-${ses_id}.nii \
-anat_has_skull no \
-anat_follower anat_w_skull anat ${T1_path} \
-blocks tshift align tlrc volreg mask blur scale regress \
-radial_correlate_blocks tcat volreg \
-tcat_remove_first_trs 0 \
-tshift_opts_ts -tpattern @${st_path} \
-align_unifize_epi local \
-align_opts_aea -cost lpc+ZZ -giant_move -check_flip \
-tlrc_base ${MNI_path} \
-tlrc_NL_warp \
-tlrc_NL_warped_dsets \
${prewarp_path}/anatQQ.sub-${sub_id}_ses-${ses_id}.nii \
${prewarp_path}/anatQQ.sub-${sub_id}_ses-${ses_id}.aff12.1D \
${prewarp_path}/anatQQ.sub-${sub_id}_ses-${ses_id}_WARP.nii \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-volreg_compute_tsnr yes \
-mask_epi_anat yes \
-blur_size 6.0 \
-regress_stim_types AM1 \
-regress_basis 'dmBLOCK' \
-regress_stim_times sub-${sub_id}_ses-${ses_id}_task-${task_id}_beh/* \
-regress_stim_labels text shape alc nonalc motor \
-regress_local_times \
-regress_opts_3dD \
-gltsym 'SYM: +alc -nonalc' \
-glt_label 1 alc_vs_nonalc \
-allzero_OK \
-GOFORIT 6 \
-jobs $cpu_num \
-regress_motion_per_run \
-regress_censor_motion 0.2 \
-regress_censor_outliers 0.05 \
-regress_3dD_stop \
-regress_reml_exec \
-regress_compute_fitts \
-regress_make_ideal_sum sum_ideal.1D \
-regress_est_blur_epits \
-regress_est_blur_errts \
-regress_run_clustsim no \
-html_review_style pythonic \
-execute
Thanks so much!
Ryann