Hi Paul,
Thank you again for answering with so much detail - it really helps when trying to work things out.
I have been trying different things with regard to what you detailed - and it seems that it's working out pretty good.
I had a couple of questions -
(1) I've been having difficulties with getting a good skull strip using SSwarper2. I've tried many variations of the command, including:
for file in anatQQ.*; do
if [[ -f $file ]]; then
echo 'anat.QQ FILES exsist'
break
else
cd $path
echo 'preforming @SSwraper'
export OMP_NUM_THREADS 4
sswarper2 \
-input anat_mask_inv2/$subject.$experiment.$session.mp2r.den_SS+orig.BRIK \
-base $abin_path/MNI152_2009_template_SSW.nii.gz \
-subid $subject.$experiment.$session.$anattype \
-odir $output_path \
-unifize_off \
-aniso_off \
-ceil_off
# -init_skullstr_off \
fi
done
No matter how hard I push the SS to 'do as less as you can' I still sometimes miss some parts of cerebellum and temporal lobe.
I've opted instead to use 3dSkullstrip directly with the Inv-2 as a base - and it seems to get pretty good results. I then use that result as a mask for the rest of my mp2rage sequences, including the denoised one, which is the dset I'm trying to move to MNI.
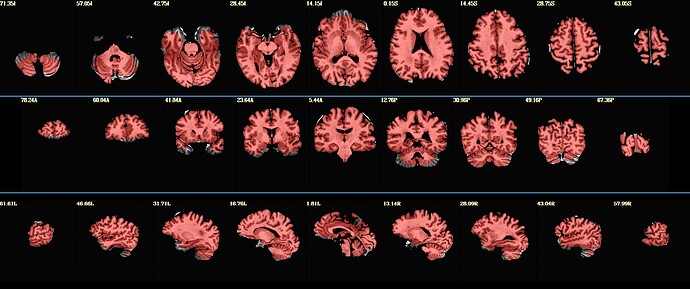
As you can see in the code above - I'm inputting a SS mp2rage denoised dataset to move to MNI (mp2r.den=mp2rage denoised), but I can't seem to find an option to let SSwarper know I'm only interested in the transition to template, without the stripping. See attached anatSS file -
I also tried using the mask_ss option -
sswarper2 \
-input anat_mask_inv2/$subject.$experiment.$session.mp2r.den_SS+orig.BRIK \
-base $abin_path/MNI152_2009_template_SSW.nii.gz \
-subid $subject.$experiment.$session.$anattype \
-odir $output_path \
-mask_ss $data/7TMS/$subject/anat/$session/mp2r.inv2/anat_mask_inv2/Automask_inv2+orig.BRIK
fi
The result from this looks awfully similar to the one I posted above.
I then tried running again using SSwarper (#1, not 2) -
@sswarper \
-input anat_mask_inv2/$subject.$experiment.$session.mp2r.den_SS+orig.BRIK \
-base $abin_path/MNI152_2009_template_SSW.nii.gz \
-subid $subject.$experiment.$session.$anattype \
-odir $output_path \
-mask_ss $data/7TMS/$subject/anat/$session/mp2r.inv2/anat_mask_inv2/Automask_inv2+orig.BRIK
fi
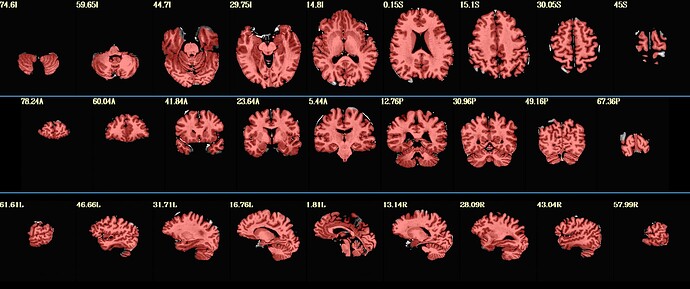
The result looks a little better, but still not perfect (or is it good enough? I'm not sure - I'm posting the photo in the next post b/c of the media limit) -
I also tried running the init_skullstr_off option (although I'm not sure it fits here) and it caused an error saying that the option is not known..
The thing is that the SS using 3dSkullstrip outside of SSwarper looks really good! I also tried stripping with Freesurfer and that also looks good.
So it boils down to several options (I think  )
)
a. Using SSwarper without any stripping at all - is there a way to do so?
b. Using another AFNI command to move to MNI, and receiving the 2 warp files that SSwarper outputs in order to use them as a matrix for the other anatomical sequences (FLAIR, SWI, T2*, T2).
Which commands should I look at? How should I use them?
The issue with option b is that I think that might cause problems when I move on to using the proc.py command, as SSwarper outputs nice files that proc needs and is sometimes upset otherwise :). If I move to MNI using a dataset that I stripped with Freesurfer, how can I make sure that I have everything I need to use proc py later on?
I hope this is clear, Thank you again!!!!