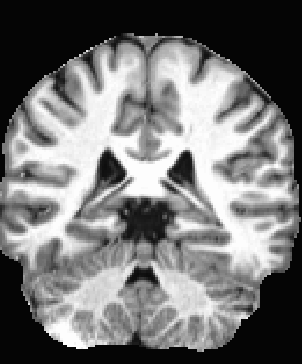
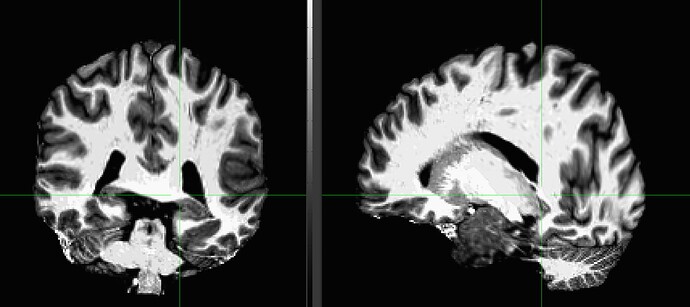
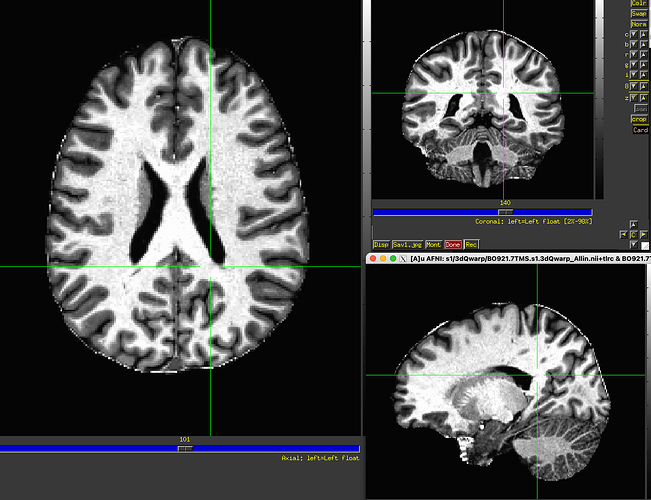
AFNI version info (afni -ver): 24.1
Hello Afni Gurus!
Our lab uses proc.py to preprocess ME data. We use an MP2RAGE sequence for registration.
Our Pipeline is comprised of the following general steps -
(1) Dicom to AFNI
(2) Creating an anatomical mask using the INV2 series acquired during MP2RAGE, and using that mask for skull stripping:
- 3dUnifize with INV2 as an input
- 3dWarp -deoblique with both INV2 and the unified series from MP2RAGE
- 3dresample post deobliquing
- 3dskullstrip post 3dresample --> creating SSanat2
- 3dAutomask --> 3dcalc to create mask
I'm attaching this bit of code here for clarity -
#### correction for the inhomogeneity of the field to file anat2
3dUnifize -prefix ${subject}.7TMS.${session}.anat2.uni $path_session/'anat2'/${subject}.7TMS.${session}.anat2+orig.
#### Deoblique for files anat 2 (corrected) and anat
3dWarp -deoblique -prefix ${subject}.7TMS.${session}.anat2.Deobl ${subject}.7TMS.${session}.anat2.uni+orig
3dWarp -deoblique -prefix ${subject}.7TMS.${session}.anat.Deobl $path_session/'anat'/${subject}.7TMS.${session}.anat+orig.
#### correct the orientation (that is changed when deoblique)
3dresample -orient ASR -rmode NN -prefix ${subject}.7TMS.${session}.anat2.DeoblASR -input ${subject}.7TMS.${session}.anat2.Deobl+orig.
3dresample -orient ASR -rmode NN -prefix ${subject}.7TMS.${session}.anat.DeoblASR -input ${subject}.7TMS.${session}.anat.Deobl+orig.
#### 3dskullstrip for anat2 and create mask
3dSkullStrip -input ${subject}.7TMS.${session}.anat2.DeoblASR+orig -prefix SSanat2
3dAutomask -prefix Automask SSanat2+orig
#### create anat mask out of the automask
3dcalc -a Automask+orig -b ${subject}.7TMS.${session}.anat.DeoblASR+orig -expr 'a*b' -prefix ${subject}.7TMS.${session}.anatMASK
Following these steps we use proc.py. We use SSwarper to create the inputs proc.py requires in order to run (anatQQ/SS). As input, SSwarper receives the final product of the bit of code pasted above - which is a skull-stripped MP2RAGE image. We are aware of the fact that SSwarper requires a non-skull stripped input, but unfortunately it isn't doing a good job with stripping our original MP2RAGE image :(.
This is the bit of code that follows -
cd $path
for file in anatQQ.*; do
if [[ -f $file ]]; then
echo 'anat.QQ FILES exsist'
break
else
echo 'preforming @SSwraper'
export OMP_NUM_THREADS 4
@SSwarper \
-input $subject.$experiment.$session.anatMASK+orig.BRIK \
-base $abin_path/MNI152_2009_template_SSW.nii.gz \
-subid $subject.$experiment.$session
fi
done
for scan in ${scans[@]}
do
echo "preforming proc.py to scan $scan"
afni_proc.py \
-subj_id $subject.$experiment.$session.$scan \
-out_dir ${output_path}/$subject.$experiment.$session.$scan.results \
-script ${output_path}/proc.$subject.$experiment.$session.$scan \
-blocks despike tshift align tlrc volreg mask combine scale regress \
-radial_correlate_blocks tcat volreg \
-blip_forward_dset $subject.$experiment.$session.AP+orig.HEAD -align_unifize_epi local \
-blip_reverse_dset $subject.$experiment.$session.PA+orig.HEAD \
-copy_anat anatSS.$subject.$experiment.$session.nii \
-anat_has_skull no \
-dsets_me_run $subject.$experiment.$session.${scan}*0?+orig.HEAD \
-echo_times 13.2 34.72 56.24 \
-reg_echo 1 \
-tcat_remove_first_trs 4 \
-tshift_interp -wsinc9 \
-align_opts_aea -cost lpc+ZZ -check_flip \
-tlrc_base MNI152_2009_template_SSW.nii.gz \
-tlrc_NL_warp \
-tlrc_NL_warped_dsets anatQQ.$subject.$experiment.$session.nii anatQQ.$subject.$experiment.$session.aff12.1D anatQQ.$subject.$experiment.${session}_WARP.nii \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-mask_epi_anat yes \
-mask_apply anat \
-combine_method m_tedana \
-html_review_style pythonic \
-scr_overwrite
tcsh -xef $output_path/proc.$subject.$experiment.$session.$scan 2>&1 | tee $output_path/output.proc.$subject.$experiment.$session.$scan
## copy final output to shared folder
echo "copying final errts and anat files to shared folder: $shared_output"
cp ${output_path}/$subject.$experiment.$session.$scan.results/errts* $shared_output
done
cp ${output_path}/$subject.$experiment.$session.$scan.results/anat_final* $shared_output/anat_final.$subject.$experiment.$session
done
done
The problem is that no matter what options we try, SSwarper (and also SSwarper2) sometimes takes a bite out of the skull stripped input it receives, taking out part of the cortex / cerebellum.
For instance -
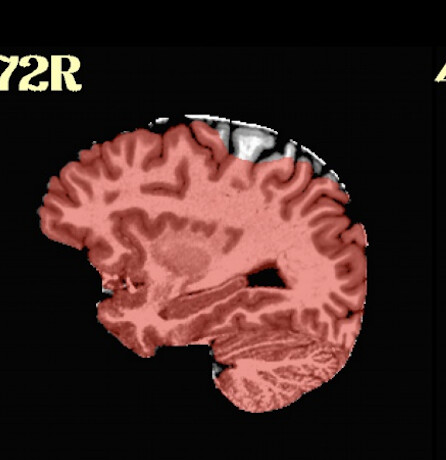
Any suggestions on how we can work this out? We have a method for skull stripping without SSwarper. We want to move to MNI, and use that product with proc.py, but to avoid skull stripping any further.
- Is there a way to add a flag to SSwarper to prevent it from Skull Stripping, and just warping to MNI?
- Any way creating the files proc.py needs without using SSwarper?
Thank you in advance!!!!!