\
Hi there AFNI wizards!
I’ve recently gotten some nice success with preprocessing of some T2-weighted EPI sequence functional datasets through afni_proc.py. The method of alignment worked best was “-align_opts_aea -cost lpa -big_move”, though I think there is room for improvement. I’ve also tried lpc+zz and nmi (both with and without -big_move), but these methods didn’t work as well as lpa -big_move.
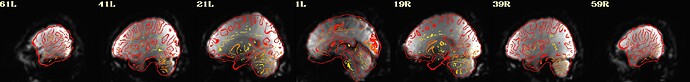
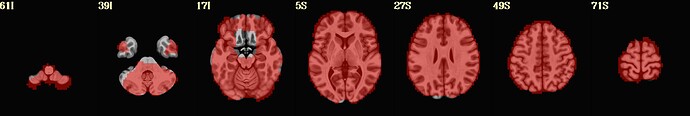
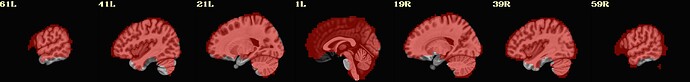
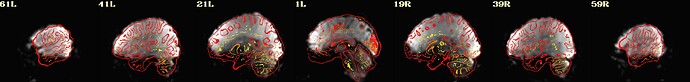
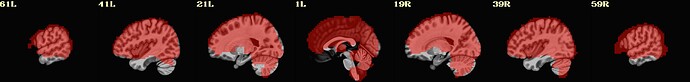
I include here some of the QC images that I think are most revealing, all from the same subject. 3 of them are from 1 scan and 3 from the other. The “meditation” scan seems to be processed in a superior manner compared to the “rest”, and I’m uncertain why. I know that the 2 datasets have slightly different TRs and I wonder if this could cause this difference in pre-processing success.
Is there any guidance you can provide about whether the results specific to the “rest” condition can be improved?
The afni_proc.py command for these data sets looks like this:
afni_proc.py \
-subj_id sub_"$1"_rest1 \
-out_dir $directory_processed/fMRI/rest1 \
-dsets $work/sub_$1/sub-"$1"_task-rest_run-01_bold.nii \
-blocks despike tshift align tlrc volreg mask blur \
regress \
-copy_anat $directory_sswarper/anatSS.sub_$1.nii \
-anat_has_skull no \
-tcat_remove_first_trs 4 \
-align_unifize_epi local \
-align_opts_aea -cost lpa \
-big_move \
-volreg_align_e2a \
-volreg_align_to MIN_OUTLIER \
-volreg_tlrc_warp \
-tlrc_base MNI152_2009_template_SSW.nii.gz \
-tlrc_NL_warp \
-tlrc_NL_warped_dsets $directory_sswarper/anatQQ.sub_$1.nii \
$directory_sswarper/anatQQ.sub_$1.aff12.1D \
$directory_sswarper/anatQQ.sub_$1_WARP.nii \
-volreg_post_vr_allin yes \
-volreg_pvra_base_index MIN_OUTLIER \
-mask_segment_anat yes \
-mask_segment_erode yes \
-regress_bandpass 0.01 0.25 \
-regress_censor_first_trs 4 \
-regress_anaticor \
-regress_ROI WMe CSFe \
-regress_apply_mot_types demean deriv \
-regress_motion_per_run \
-regress_censor_motion 0.3 \
-regress_censor_outliers 0.1 \
-blur_size 3.0 \
-regress_est_blur_epits \
-regress_est_blur_errts \
-html_review_style pythonic \
-execute