AFNI version info (afni -ver): Precompiled binary macos_13_ARM_clang: Jan 18 2024 (Version AFNI_24.0.01 'Caracalla')
Hi experts,
I am using @chauffeur to save some figures of my results. At the same time, I want to save the threshold filtered image as a mask file for further analyses.
With the C2S file I got from @chauffeur, the overlay is all red. I cannot save it as a mask (such as using the clusterize and then Rpt and then SaveMsk.
Is this function possible? Or I have to save it mannually?
My code:
@chauffeur_afni -ulay TT_N27.nii \
-olay dr_stage3_ic0027_tfce_corrp_tstat6.nii.gz \
-prefix Fig_70IC_IN_SGR27 \
-thr_olay 0.95 \
-set_xhairs OFF \
-montx 8 -monty 1 \
-delta_slices -1 -1 1 \
-opacity 8 \
-set_dicom_xyz 35 -56 35 \
-zerocolor white \
-cbar Reds_and_Blues_Inv \
-label_mode 1 \
-label_size 4 \
-label_color Black \
-no_cor \
-no_sag \
-cmd2script C2S_70IC_IN_SGR27
The results of C2S scripts:
Thank you in advance!
Best,
Xiyue
Hi, Xiyue-
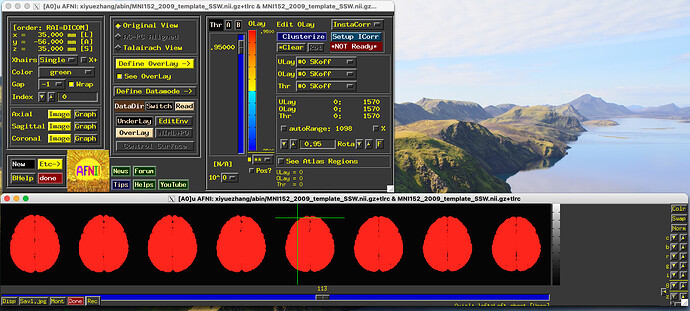
It is possible that the script would work OK, but the datasets being shown there as underlay and overlay are the MNI152_2009_template_SSW.nii.gz dset, not the desired ones in your script. I can see this in the filenames shown in the panel par containing the AFNI GUI. And indeed, the [0]th volume of that MNI*SSW.nii.gz dset is 1570, as shown in the min/max ranges for the Ulay and Olay.
So, I think the issue is that the created script cannot get the correct filenames that you would like, which are actually:
-ulay TT_N27.nii \
-olay dr_stage3_ic0027_tfce_corrp_tstat6.nii.gz \
There might be error messages in the terminal text about that, and so the GUI is using other default images it can find, instead (here, MNI*SSW.nii.gz)
--pt
Hi Paul,
Thank you for your quick reply. I added the pathway of the -ulay and -olay and the C2S works! Then I can save the mask mannully. Since I have many many masks to save, is it posible that a command can do it automatically?
Thanks!
Xiyue
Hi Paul,
I think I found it:
3dClusterize -nosum -1Dformat -inset dr_stage3_ic0027_tfce_corrp_tstat6.nii.gz -idat 0 -ithr 0 -NN 1 -clust_nvox 1 -bisided -0.95 0.95 -pref_map Msk_70IC_IN_SGR27_mask
Thanks !
Xiyue
OK, cool. And note you can run clusterize within @chauffeur_afni, too. I think you are set here already, but just so you know about the functionality for future cases:
-clusterize "-opt0 v0 -opt1 v1 ..."
:input a set of options "-opt0 v0 -opt1 v1 ..." for
3dClusterize to use internally, so that the
overlay dataset is clusterized. Can be combined
with Alpha+Boxed. See the "Clusterize
capabilities" description in the NOTES below for
what options go where when clusterizing. Examples
are also included below.
-clusterize_wami CW :if using '-clusterize ..', then this option can be used
to run AFNI's whereami program on the results. The user
provides the name of an allowed atlas for reference
(see the top of whereami's help), and a text file
reporting the relative overlap of each ROI will be
produced in the outputs.
--pt
1 Like