Hi! This is regarding FreeSurfer + @SSwarper:
We have been pulling our hair out over wierd stuff happening. Following the guide, we simply run recon-all -all where we input our orig T1:
T1=/path/T1.nii
This generates the /path/freesurfer/subX dir where we run @Make_Special_FS command which generates the dir
/path/freesurfer/subX/SUMA where we find all masks/ROIs. Also, we find a T1.nii.gz image.
Normally we procede to our afni_proc.py script, where we run @SSWarper. According to the guide we feed it with the orig T1:
@SSwarper -input FT_anat+orig
So we set:
T1=/path/T1.nii
The normal case of ordinary recon-all:
What happens is that FS resamples the input a little bit, for example it always makes the matrix “cubically-isotropic”. If the input (T1) is:
170x256x256, then the output space of e.g. wm_seg.nii is 256x256x256. This is also the dimentions of the …/SUMA/T1.nii.gz file.
The …/SUMA/T1.nii.gz file is always a perfect match with the ROIs/maks/segments.
If I would copy your orig T1 into the SUMA dir and look at how the ROIs align, it is usually fine. Even though the orig T1 does not have the same matrix dims (why is that?).
The alignment of orig T1 and the ROIs can, however, be shifted (as we noticed) from a few voxels to a lot when using the orig T1 as underlay and wm_seg.nii as overlay. This off shift does, ofc, follow into the proc.results dir when overlapping anat_final with follower_wm.nii.gz (also true when looking at copy_wm._seg.nii.gz over T1 in orig space, so not a “warping issue”) This often completely disappears if you have processed your orig T1 a little bit before feeding it into recon-all by setting the center of the image to (0,0,0) and cut out some of the neck (yes, we have been up late testing things).
For example, one subject had horrible T1 orig and wm_seg.nii aligment when just feeding it raw into FS.
But when we ran: (this is from an old discussion with Paul Taylor on old issues we had with SSWarper =)).
#get rid of space below brain
3dZeropad -I -70 -prefix sub${sub}-1_T1_zp.nii sub${sub}-1_sT1W_3D.nii
#set center of mass inside the brain
3dCM -set 0 0 0 sub${sub}-1_T1_zp.nii
And then fed FS with the zero-padded and center of mass:ed T1 the alignment was almost perfect.
What ALWAYS works though, is to just feed SSWarper with the …/freesurfer/SUMA/T1.nii.gz file instead. This simply seems to be the orig T1 but in the same space as the ROIs and the same matrix dims.
Note: I might be fooled by some de-obliqe magic here. Maybe the orig T1 is perfectly aligned with the masks/ROIs but the viewer tells me otherwise. However, the matrix dims do miss-match.
Would you recommend to use …/freesurfer/SUMA/T1.nii.gz as input to SSwarper instead of:
@SSwarper -input FT_anat+orig
Like in the documentation.
Also, this helped us:
3dZeropad -I -70 and 3dCM -set 0 0 0 on T1 prior to recon-all
I might be off here, but visually we noticed that 3dZeroPad + 3dCM on the data prior to feeding it into FS did the trick. But even with orig data without 3dCM, …/SUMA/T1.nii.gz gave really good results.
The freesurfer -longitudinal way
The guide probably assumes a normal freesurfer recon-all have been run. But sometimes, like in our dateset, you have two scans separated with a month or two. Then you can improve the quality by using the longitudinal pipeline:
Run T1.nii.gz from session 1 and T1.nii.gz from session 2 in two separate normal recon-all runs, call the output dirs TP1 and TP2.
Then run recon-all with the -base option to generate a template based on TP1 and TP2:
recon-all -base ${subject}_Template -tp TP1 -tp TP2 -all -sd $subject_dir -parallel
This gives a sub_id_Template dir
Then you run two separate longitudinal recon-all runs on the original data:
#TP1
recon-all -long TP1 ${subject}_Template -all -sd $subject_dir
#TP2
recon-all -long TP2 ${subject}_Template -all -sd $subject_dir
This gives new outout dirs:
TP1_subid.long
TP2_subid.long
I.e. segmentation but it has used the knowledge of two scans, separated in time, to get better results.
We can then run @Make_Special_FS in each of these .long-dirs to get the SUMA dir with the ROIs/masks for TP1 and TP2. We are just interested in the second scan (TP2) but use TP1 to improve segmentation.
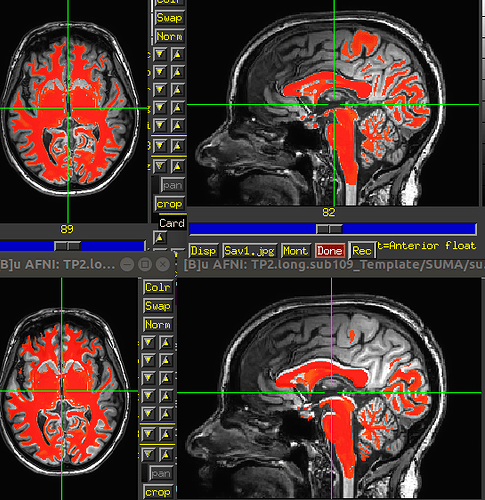
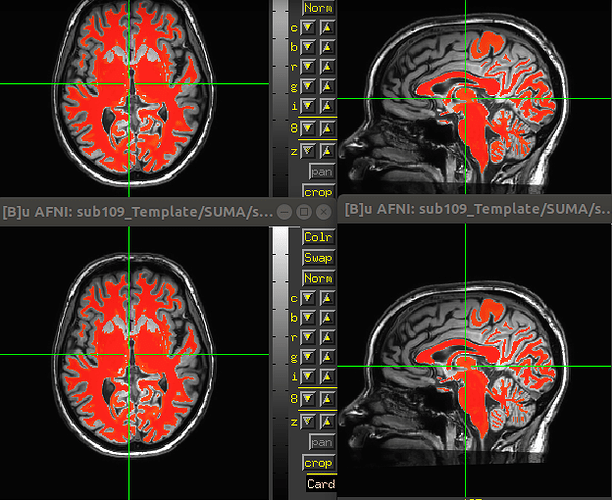
As I understand it, these ROIs are in “template space”. It is quite a big miss-aliment of the …/TP2.long/SUMA/wm_seg.nii.gz and the orig TP2 orig input image. I compared the shift to the normal single run (with bad aligment due to not using 3dCM) and the shift is different. The respective TPX.long/SUMA/T1.nii.gz file matches perfectly, again, with the ROIs. TP1.long/SUMA/T1.nii.gz and TP2.long/SUMA/T1.nii.gz are pretty much perfectly aligned which indicates that we are in a “common space” between the two orig T1 images. I.e. Template space.
The only way for us to make the ROIs match was to feed SSwarper with the TP2.long/SUMA/T1.nii.gz file instead of the orig time point 2 T1 image (TP2 since we only want the data from the second fMRI session).
Our question, does our fix sound legit in the longitudinal case? I.e., point to the T1.nii.gz file in the SUMA dir of the corresponding longitudinal time point instead of the orig data:
#we replaced
@SSwarper -input FT_anat+orig
#with
@SSwarper -input $SUMA_dir/T1.nii.gz
I am aware that I can be completely wrong, but this is the only conclusion we came up with after working on this for a couple of days.
This was done in collaboration with another AFNI-user, Irene Perini =)
Please let me know if we made a huge mistake somewhere or if we could be onto something.
Thanks!