Dear AFNI experts,
I have processed EPI data as shown in the afni_proc.py code below.
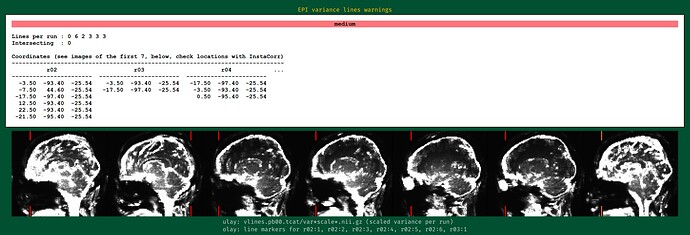
When I checked the results with QC, I got the EPI line variance warning as shown in the image.
In previous discussions, it was suggested that this EPI variance could be an issue with the MRI vendor. (The MRI I am using is a Siemens Magnetom Prisma 3T MRI).
My question is, can the subject's motion also cause these warnings?
The reason I came to this question is that when I processed data from subjects who performed the same task, I noticed differences in simple GLM results such as the move-rest contrast for each subject.
Also, I would like to get advice on whether these issues can be resolved by modifying the current preprocessing code, and if so, what modifications should be noted.
Thank you for your kind advice,
Yonghyun
AFNI version info (
afni -ver): Precompiled binary linux_ubuntu_16_64: Mar 24 2024 (Version AFNI_24.0.17 'Caracalla')
#!/bin/zsh
## zsh s2-ii---.zsh -s (Pilot??) -d (1 or 2)
## ======================================= ##
## $# = the number of arguments
while (( $# )); do
key="$1"
case $key in
## pattern)
## sentence
## ;;
-s | --subject)
subj="$2"
;;
-d | --day)
day="$2"
;;
esac
shift ## takes one argument
done
echo $subj $day
## ======================================= ##
dir_root="/mnt/ext2/yonghyun/GAB_project/GAB_2024/fMRI_data"
dir_cvt="$dir_root/cvt_data/$subj"
dir_mask="$dir_root/mask_data"
## ======================================= ##
## I/O path, same as above, following earlier steps
dir_FreeSurfer="$dir_root/FreeSurfer"
if [[ ! -d $dir_Freesurfer ]]; then
mkdir -p -m 755 $dir_FreeSurfer
fi
if [[ ! -d $dir_FreeSurfer/$subj ]]; then
source /usr/local/freesurfer/7.2.0/SetUpFreeSurfer.sh
export SUBJECTS_DIR=${dir_FreeSurfer}
## FS function
recon-all \
-sid $subj \
-sd $dir_FreeSurfer \
-i $dir_cvt/T1.$subj.nii \
-all \
## AFNI-SUMA function: convert FS output
@SUMA_Make_Spec_FS \
-NIFTI \
-fspath $dir_FreeSurfer/$subj \
-sid $subj
3dQwarp -allineate -blur 0 3 \
-base "/usr/local/afni/abin/MNI152_2009_template_SSW.nii.gz" \
-source "$dir_FreeSurfer/$subj/SUMA/brain.nii.gz" \
-prefix "$dir_FreeSurfer/$subj/SUMA/brain_qw.nii"
fi
## ======================================= ##
dir_output="$dir_root/proc_data/${subj}_${day}"
dir_script="/home/yonghyun/Ongoing/GAB_project/preprocessing/GAB2_afni_proc.py"
dsets=(`find $dir_cvt -type f -name "func.$day.r0?.$subj.nii" | sort -t ' ' -k 1`)
echo $dsets
cd $dir_script
afni_proc.py \
-subj_id $subj \
-out_dir $dir_output \
-blocks despike tshift align tlrc volreg blur mask scale regress \
-radial_correlate_blocks tcat volreg \
-copy_anat "$dir_FreeSurfer/$subj/SUMA/brain.nii.gz" \
-anat_has_skull no \
-anat_follower anat_w_skull anat "$dir_FreeSurfer/$subj/SUMA/T1.nii.gz" \
-anat_follower_ROI aaseg anat \
"$dir_FreeSurfer/$subj/SUMA/aparc.a2009s+aseg_REN_all.nii.gz" \
-anat_follower_ROI aeseg epi \
"$dir_FreeSurfer/$subj/SUMA/aparc.a2009s+aseg_REN_all.nii.gz" \
-anat_follower_ROI FSvent epi "$dir_FreeSurfer/$subj/SUMA/fs_ap_latvent.nii.gz" \
-anat_follower_ROI FSWe epi "$dir_FreeSurfer/$subj/SUMA/fs_ap_wm.nii.gz" \
-anat_follower_erode FSvent FSWe \
-dsets $dsets \
-tcat_remove_first_trs 0 \
-blip_forward_dset $dir_cvt/func.$day.r01.$subj.nii \
-blip_reverse_dset $dir_cvt/func.$day.reverse.$subj.nii \
-align_unifize_epi yes \
-align_opts_aea \
-cost lpc+ZZ \
-giant_move \
-resample off \
-check_flip \
-tlrc_base MNI152_2009_template_SSW.nii.gz \
-tlrc_NL_warp \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-volreg_warp_dxyz 2.0 \
-blur_size 4 \
-mask_epi_anat yes \
-regress_motion_per_run \
-regress_ROI_PC FSvent 3 \
-regress_ROI_PC_per_run FSvent \
-regress_make_corr_vols aeseg FSvent \
-regress_anaticor_fast \
-regress_anaticor_label FSWe \
-regress_censor_motion 0.4 \
-regress_censor_outliers 0.05 \
-regress_apply_mot_types demean deriv \
-html_review_style pythonic