AFNI version info Precompiled binary linux_openmp_64: May 4 2023 (Version AFNI_23.1.02 'Publius Helvius Pertinax')
Hej AFNI Gods, long time no see.
We have developed our own neurofeedback engine and I end up with a problem.
All processing is done in .orig space. We run a localizor, create a mask and then we align/tranform that mask onto the current epi.orig volume. Which works fine.
Now that the runs are done I have a T1.orig file that is pretty well overlapping with the stats-file, that we used to calculate our mask. We also run a carry-over scan at the end of the three sessions long neuro-feedback treatment study.
Now, I would like to compare these stats files, one collected at the first session (in orig space) and one that was create in the last session (in that orig space).
My idea was simply to warp the T1 to MNI space and then use those warping parameters to warp the stats-file to MNI space. First for the localizor scan and then for the carry over scan, on order to be able to compare the stats_localizor_mni map with the carryover_mni map in 3dMVM.
I tried this:
auto_warp.py -base MNI152_2009_template_SSW.nii.gz -input anat_final.ref+orig. -followers stats.ref+orig.
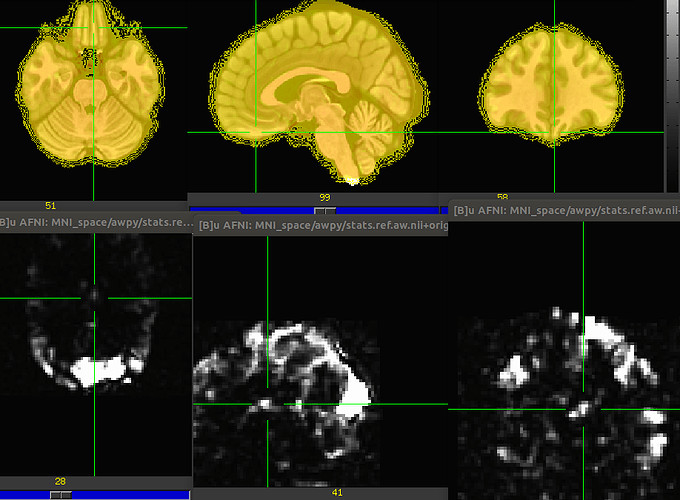
Which gives me a nice overlap between the template and the anatomical scan but the stats-map is all wrong. I am attaching a print screen where the top row is base.nii (MNI template) overlayed with the anat_final_ref_aw and the bottom row is a new afni window with stats.ref.aw.
Is there a way to take a T1.orig map, with a matched (at least visually) stats-map to MNI space?