Hi, Congcong-
No need to apologize for not working 24-7! And thanks for posting that.
I guess from the number of CPUs on that computer, and the path location of your AFNI binaries, that that might be some kind of shared, high-performance computer? If so, a solution might involve asking your IT support there to install a couple more dependencies.
Firstly, you can run the following, that are suggested at the bottom of the system check output in the "please fix" section:
cp /opt/fox_cloud/share/app/imaging/afni/AFNI.afnirc ~/.afnirc
suma -update_env
apsearch -update_all_afni_help
That will populate your default AFNI environment variables and SUMA environment variables, and then setup your system to autocomplete options when typing AFNI commands in the terminal.
Next, your AFNI version is now over 3 years old---I would strongly advise you to update it (or ask your IT to update it). If they cannot do so, you actually could download a set of precompiled binaries under your own directory, and add that location to your path.
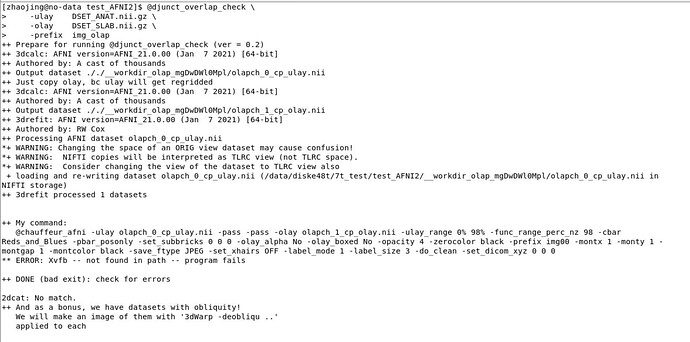
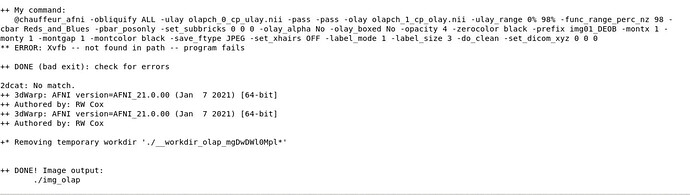
But that will likely still require updating some dependencies. In more modern AFNI versions, we check for more dependencies and report that in the afni_system_check.py output. The list of dependencies for CentOS is here. Perhaps you could share that with your IT, particularly this section? One that is definitely missing on that system is Xvfv (which is installed by that CentOS package manage as xorg-x11-server-Xvfb, as shown in that list of dependencies).
Would you be able to try that, and then we can work on the data question itself?
--pt