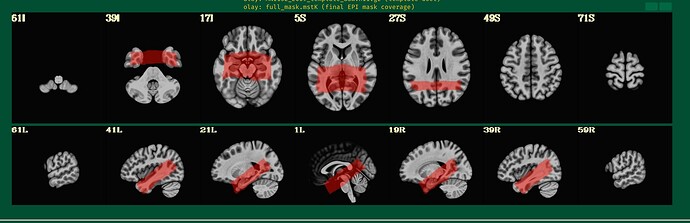
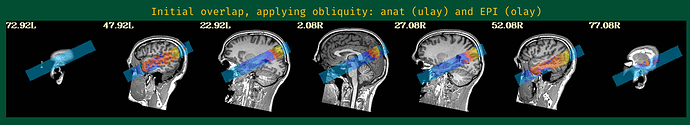
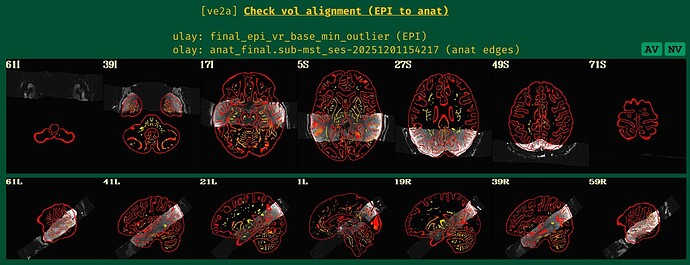
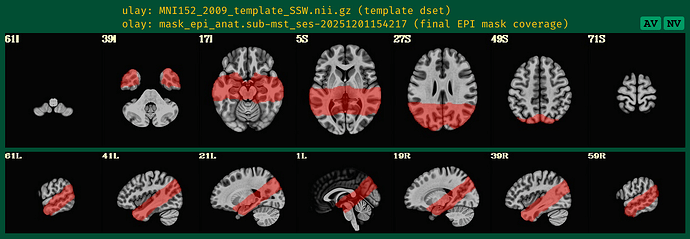
AFNI version info (afni -ver):
Precompiled binary linux_ubuntu_16_64: May 23 2025 (Version AFNI_25.1.11 'Maximinus')
Dear AFNI,
I am following up on the forum thread partial coverage alignment failure from January 2024 that seemed to stop because of technical issues. I am having a very similar issue since I am also trying to acquire a few BOLD fMRI slices including the hippocampus.I have only tried afni_proc so far, both on MNI (i.e. with tlrc option) and on subject space (ithout tlrc option). Unless you think it would be better to focus on MNI script, I will focus on T1w space since I am running only one participant to tune the parameters for now.
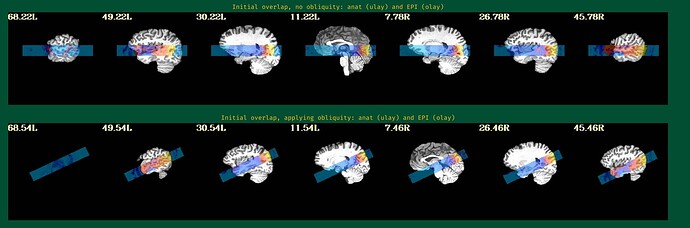
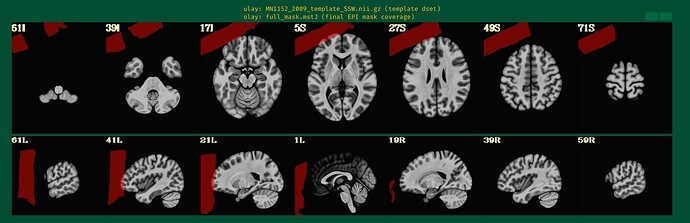
This is the beginning of output. When checking the volumes in original space, the images when applying obliquity look almost correct. That's what I acquired.
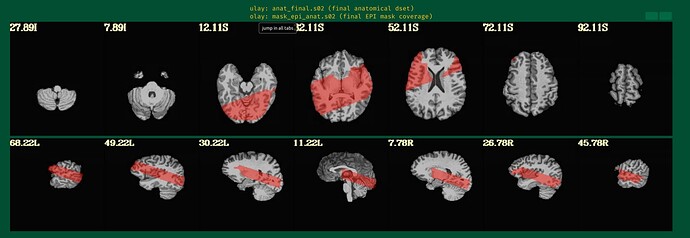
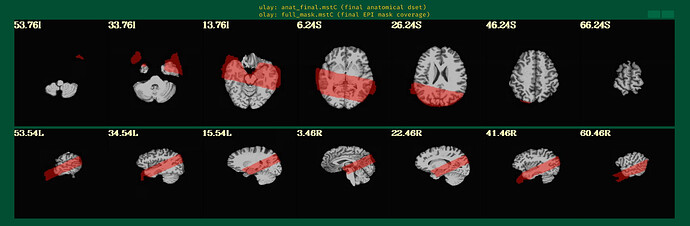
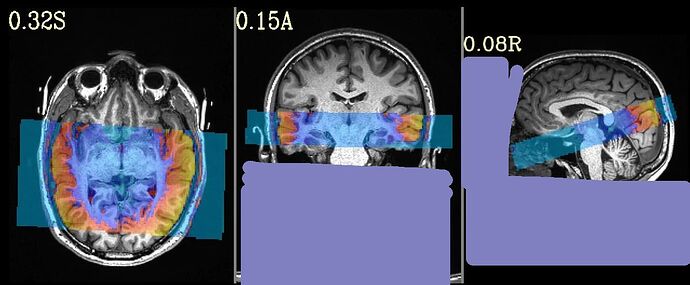
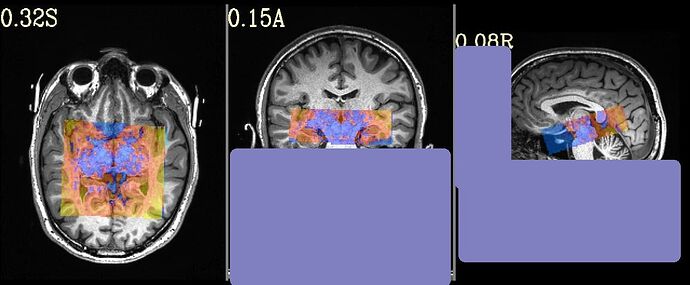
But when checking for EPI to Anat alignment, it looks like follows (almost like applying re-obliquing twice):
As an answer to the previous thread I am following, this is the output of
3dinfo -obliquity -prefix anat_final.s02+orig final_epi_vr_base_min_outlier+orig
9.366 anat_final.s02
0.000 final_epi_vr_base_min_outlier
and
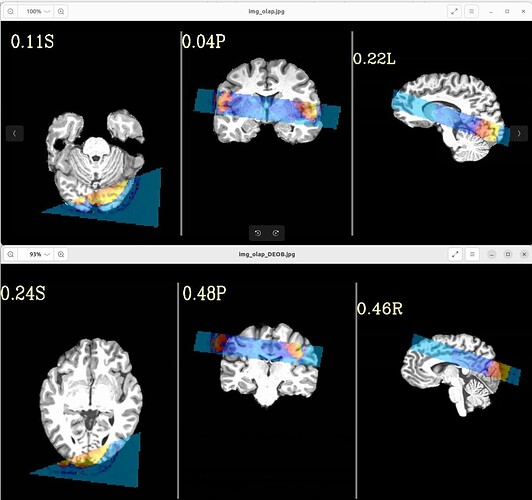
@djunct_overlap_check \
-ulay anat_final.s02+orig \
-olay final_epi_vr_base_min_outlier+orig \
-prefix img_olap
returned
# ulay = anat_final.s02+orig
# ulay_is_obl = 1
# ulay_obl_ang = 9.366
# mat44 Obliquity Transformation ::
0.995392 0.007651 -0.095584 -11.890205
-0.020267 0.991080 -0.131722 -12.362091
0.093724 0.133052 0.986668 -28.940544
#
# olay = final_epi_vr_base_min_outlier+orig
# olay_is_obl = 1
# olay_obl_ang = 9.366
# mat44 Obliquity Transformation ::
1.000000 0.000000 0.000000 -0.000008
0.000000 1.000000 0.000000 0.000000
0.000000 0.000000 1.000000 0.000000
with following images:
Where the DEOB doesn't seem to be correct.
Is there a possibility to include a full brain volume (which I acquired on many more slices and totally aligned with the time series focused on hippocampus) in the afni_proc command or have I done any mistake in my command which follows?
afni_proc.py \
-subj_id s02 \
-out_dir $subdir/S02e.results \
-copy_anat $subdir/o.ssw_sub-mst/anatSS.sub-mst.nii \
-anat_has_skull no \
-anat_follower anat_w_skull anat $subdir/sub-mst_T1w.nii.gz \
-dsets $subdir/sub-mst_task-cmrr_run-01_bold.nii.gz \
-blocks align volreg mask blur \
scale regress \
-radial_correlate_blocks tcat volreg \
-tcat_remove_first_trs 0 \
-align_unifize_epi local \
-align_opts_aea -cost lpc+ZZ \
-giant_move \
-check_flip \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_compute_tsnr yes \
-mask_epi_anat yes \
-blur_size 3.0 \
-regress_stim_times $subdir/rep.1D \
$subdir/new.1D \
-regress_stim_labels rep new \
-regress_basis_multi 'TENT(0,16,7)' 'TENT(0,16,7)' \
-regress_opts_3dD -jobs 8 \
-gltsym 'SYM: rep - new' \
-glt_label 1 r-n \
-regress_motion_per_run \
-regress_censor_motion 0.3 \
-regress_censor_outliers 0.05 \
-regress_3dD_stop \
-regress_reml_exec \
-regress_compute_fitts \
-regress_make_ideal_sum sum_ideal.1D \
-regress_est_blur_epits \
-regress_est_blur_errts \
-regress_run_clustsim no \
-html_review_style pythonic \
-execute
Thanks!
Jonathan