Hi,
I want to build my specific template using SPM, which need to provide Tissue Segmentation Masks (GM, WM, CSF). I looked at https://afni.nimh.nih.gov/pub/dist/doc/htmldoc/nonhuman/macaque_tempatl/template_nmtv2.html, however, I did not find Tissue Segmentation Masks with NMT v2. So, should I choose to use other templates (e.g., NMT v1)?
Thanks.
Hi, Ruilin-
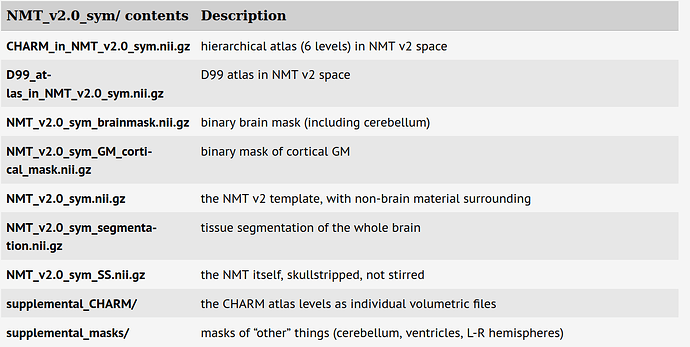
In the second table shown there:
... the 6th row of data file names is:
NMT_v2.0_sym_segmentation.nii.gz tissue segmentation of the whole brain.
That should be the file you want.
Running 3dinfo, we can see what integer value is for each type (standard WM, GM, CSF; and scGM = subcortical gray matter, and BV = arterial blood vasculature).
$ 3dinfo -labeltable ./NMT_v2.0_asym_05mm_segmentation.nii.gz
<VALUE_LABEL_DTABLE
ni_type="2*String"
ni_dimen="5" >
"4" "WM"
"5" "BV"
"1" "CSF"
"2" "GM"
"3" "scGM"
</VALUE_LABEL_DTABLE>
--pt
Hi, ptaylor
When I extracted different values from NMT_v2.0_asym_05mm_segmentation.nii.gz, I obtained some tissue masks, such as WM mask, cortical GM mask.
However, what I need are tisue probablity maps, including GM, WM, and CSF probablity. I found them from NMT v1, not NMT v2.
Hi, Ruilin-
Ah, I see. I should have paid more close attention to the title of this thread...
@dglen will be able to provide more details on whether probabilistic tissue maps are available.
There were a lot of changes between NMT v1.3 and 2.*. I think it would be highly preferable to stay in the realm of v2.*. See here for a discussion of updates/changes:
- Jung B, Taylor PA, Seidlitz PA, Sponheim C, Perkins P, Ungerleider LG, Glen DR, Messinger A (2021). A Comprehensive Macaque FMRI Pipeline and Hierarchical Atlas. NeuroImage 235:117997.
A comprehensive macaque fMRI pipeline and hierarchical atlas - PubMed
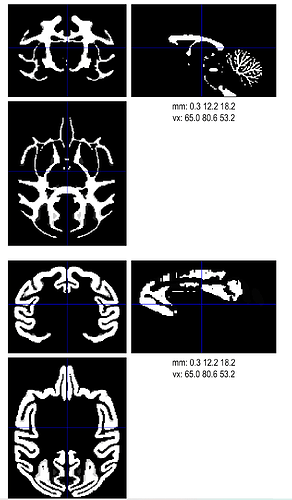
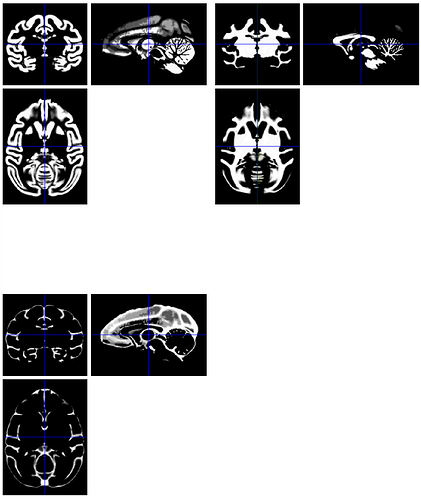
I'm a bit curious why probabilistic tissue maps would be necessary? And do they have to be exactly derived probabilities? Would you be able to take the existing binary segmentation maps, and blur them slightly for practical usage? Consider:
# make a binary mask of tissue (must be float type, for desired blurability)
3dcalc \
-a NMT_v2.1_asym_05mm_segmentation.nii.gz"<GM>" \
-expr 'step(a)' \
-prefix nmt_v2_gm.nii.gz \
-datum float \
-nscale
# blur by desired amount, like 1mm here
3dmerge \
-1blur_fwhm 1 \
-doall \
-prefix nmt_v2_gm_blur.nii.gz \
nmt_v2_gm.nii.gz
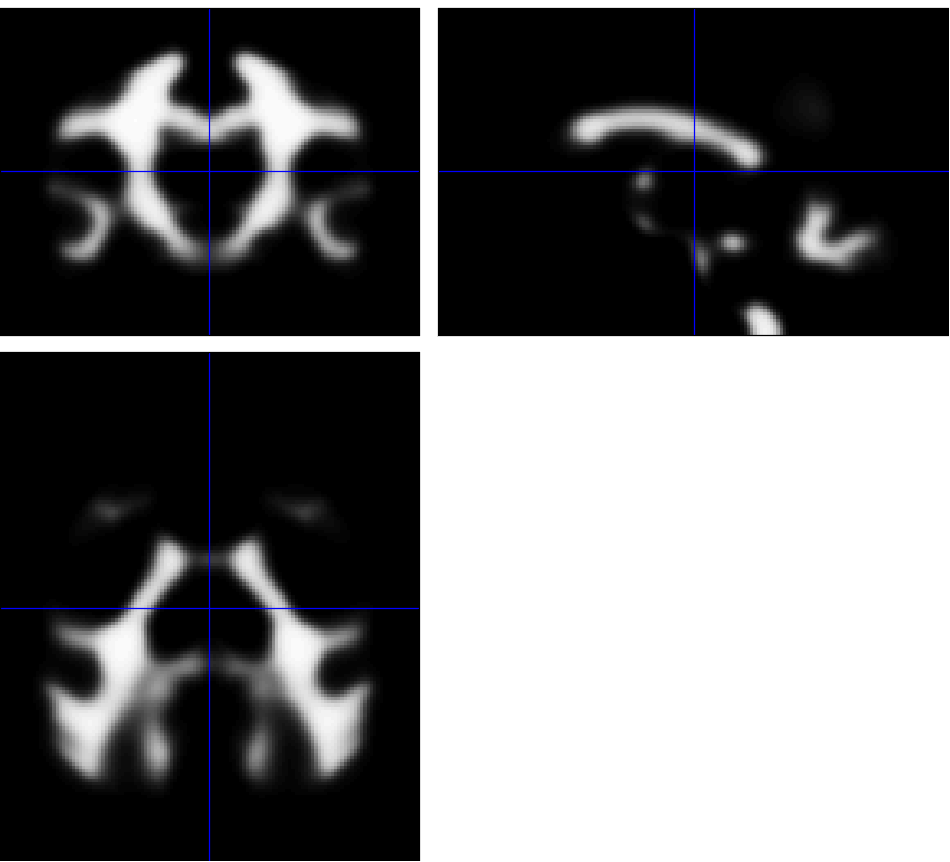
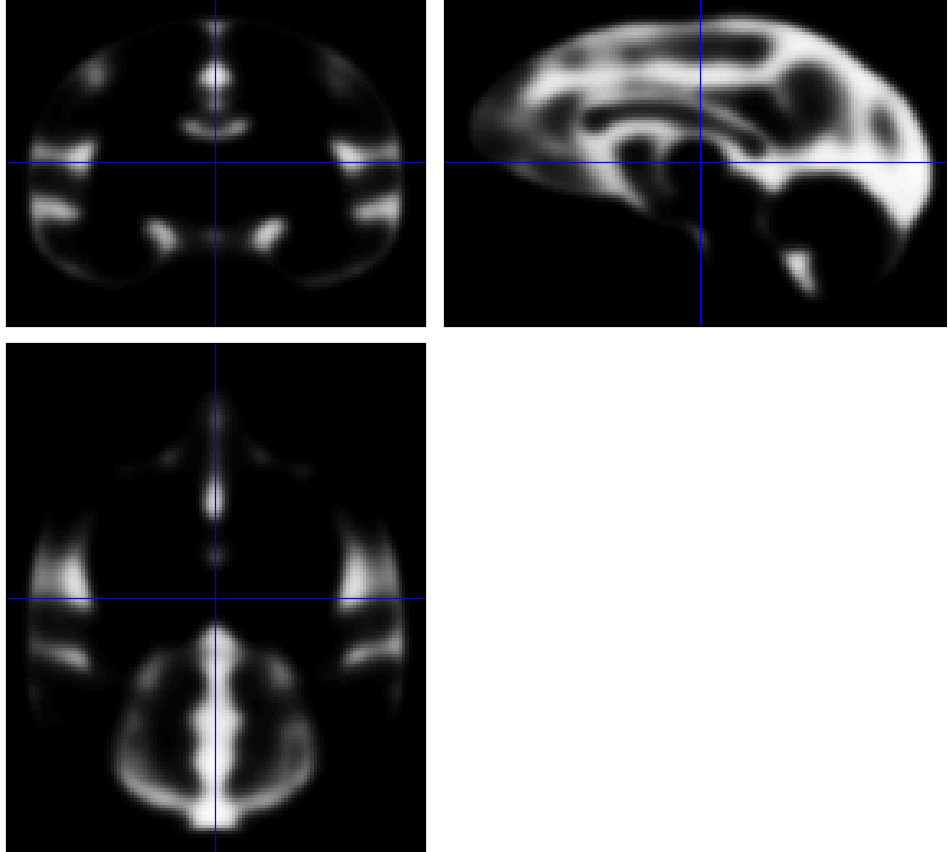
... and compare the original binarized map (top) and blurred pseudo-probability map (bottom):
Would that be usable?
--pt
I don't have the NMT 2 probabilistic maps, but I'll see if I can track them down if that's important to you. In the meantime, you could warp the maps from NMTv1.3 space to the NMT v2 template you need by getting the affine 1D transformation file and the nonlinear warp file from here:
Apply the warp to the dataset with this command (here for NMTv2 symmetric):
3dNwarpApply -iwarp \
-nwarp 'NMTv2sym_shft_WARP.nii.gz NMTv2sym_composite_linear_to_template.1D' \
-source NMT_segmentation_GM.nii.gz \
-prefix NMT_segmentation_GM_in_NMT2.nii.gz \
-master NMTv2sym.nii.gz
Alternatively, do your analysis with the NMT 1.3 version and move the results into the NMT 2 space if that is important to you.
Hi, ptaylor
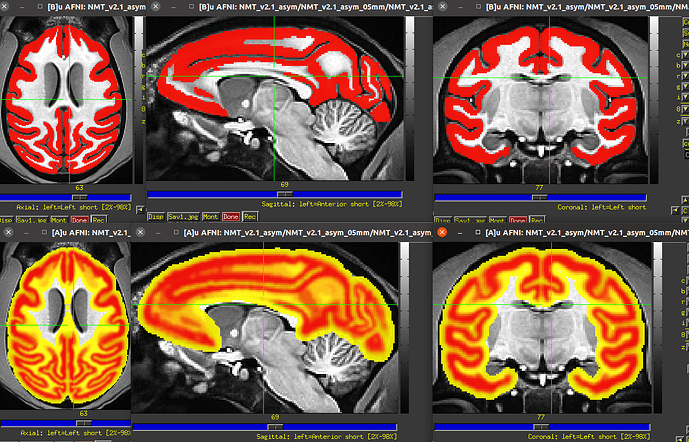
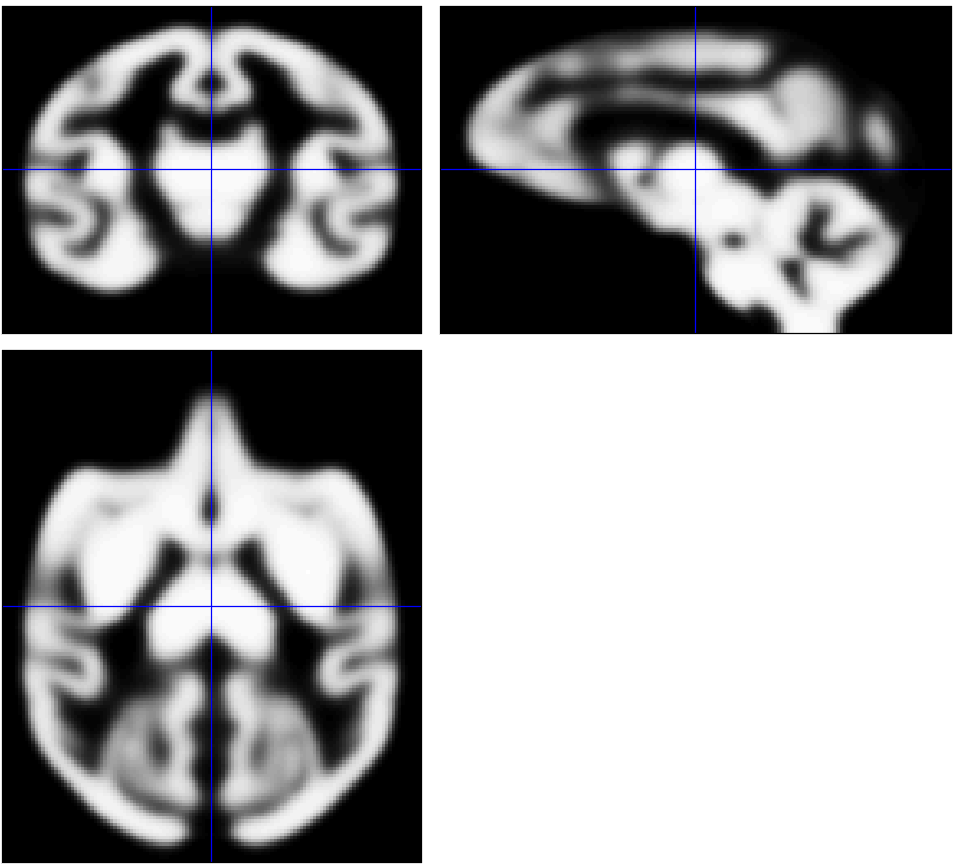
Thanks for your suggestions. I used the above code to obtain probabilistic tissue maps and then tried to create my specific template. Fortunately, I can run Dartel (create Templates) in SPM successfully. Here is my specific template (probabilistic tissue maps for GM, WM, and CSF) which used two subjects' T1 image for the purpose of testing.
Best,
Ruilin
Hi,dglen
With the aim to analyze how gray and white matter volume change with age, we wanted to create different specific age templates using Dartel tools from SPM, which need to provide tisue probablity maps. I will follow your suggestions to warp the maps from NMTv1.3 space to the NMT v2 template.
Best,
Ruilin