Hey Paul,
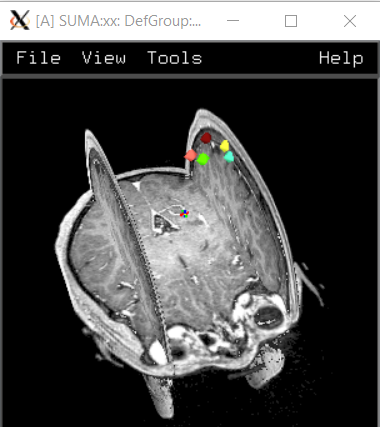
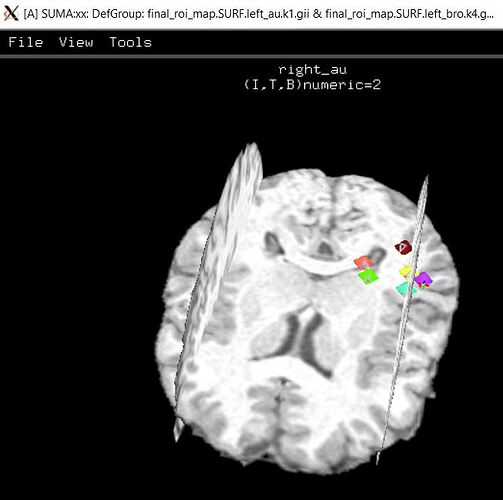
I implemented your suggestions, and am still having the same issues with ROI locations on the Suma image. On the upside, at least its generating montage images. When I right-click on all the ROI in the SUMA representation it gives more info which I’m including here along with the command line info. Let me know if you'd also like to see output from all the programs do_0?_* Any ideas on what more I can try to fix our ROI spacing issue? Thanks a lot for your time and your help.
Variable settings
set ilist = 00_list_of_all_roi_centers.txt
set vepi = all_runs.masters02-baseline+tlrc
set vanat = anat_final.masters02-baseline+tlrc.HEAD
Output from right-clicking the SUMA ROI
sandra@DESKTOP-RJJ49UB:~/data/Data/automation-test/newcases/masters02.rois/afni_tutorial_rois_sswarper_fixed_ROIs$ ./do_
00_setup_example.tcsh
++ 3dUndump: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Starting to fill via -xyz coordinates
++ Total number of voxels filled = 7
++ Wrote out dataset ./roi_mask_001.nii.gz
++ 3dUndump: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Starting to fill via -xyz coordinates
++ Total number of voxels filled = 7
++ Wrote out dataset ./roi_mask_002.nii.gz
++ 3dUndump: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Starting to fill via -xyz coordinates
++ Total number of voxels filled = 7
++ Wrote out dataset ./roi_mask_003.nii.gz
++ 3dUndump: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Starting to fill via -xyz coordinates
++ Total number of voxels filled = 7
++ Wrote out dataset ./roi_mask_004.nii.gz
++ 3dUndump: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Starting to fill via -xyz coordinates
++ Total number of voxels filled = 7
++ Wrote out dataset ./roi_mask_005.nii.gz
++ 3dUndump: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Starting to fill via -xyz coordinates
++ Total number of voxels filled = 7
++ Wrote out dataset ./roi_mask_006.nii.gz
++ Skull-stripping the anat vol.
-- Error SUMA_BrainWrap_ParseInput (SUMA_3dSkullStrip.c:1296):
Output dset ./all_runs_mprage_ss.nii.gz+tlrc exists, will not overwrite
++ DONE!
sandra@DESKTOP-RJJ49UB:~/data/Data/automation-test/newcases/masters02.rois/afni_tutorial_rois_sswarper_fixed_ROIs$ rm mprage
sandra@DESKTOP-RJJ49UB:~/data/Data/automation-test/newcases/masters02.rois/afni_tutorial_rois_sswarper_fixed_ROIs$ ./do_00_setup_example.tcsh
++ 3dUndump: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Starting to fill via -xyz coordinates
++ Total number of voxels filled = 7
++ Wrote out dataset ./roi_mask_001.nii.gz
++ 3dUndump: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Starting to fill via -xyz coordinates
++ Total number of voxels filled = 7
++ Wrote out dataset ./roi_mask_002.nii.gz
++ 3dUndump: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Starting to fill via -xyz coordinates
++ Total number of voxels filled = 7
++ Wrote out dataset ./roi_mask_003.nii.gz
++ 3dUndump: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Starting to fill via -xyz coordinates
++ Total number of voxels filled = 7
++ Wrote out dataset ./roi_mask_004.nii.gz
++ 3dUndump: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Starting to fill via -xyz coordinates
++ Total number of voxels filled = 7
++ Wrote out dataset ./roi_mask_005.nii.gz
++ 3dUndump: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Starting to fill via -xyz coordinates
++ Total number of voxels filled = 7
++ Wrote out dataset ./roi_mask_006.nii.gz
++ Skull-stripping the anat vol.
++ DONE!
sandra@DESKTOP-RJJ49UB:~/data/Data/automation-test/newcases/masters02.rois/afni_tutorial_rois_sswarper_fixed_ROIs$ ./do_01_make_single_roi_map.tcsh
++ 3dMean: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ OK: ROI masks don't appear to overlap
++ 3dTcat: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
*+ WARNING: Set TR of output dataset to 1.0 s
++ elapsed time = 0.0 s
++ 3dTstat: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./final_roi_map.nii.gz
++ 3drefit: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset final_roi_map.nii.gz
++ 3drefit processed 1 datasets
++ My command:
@chauffeur_afni -ulay anat_final.masters02-baseline+tlrc.HEAD -olay final_roi_map.nii.gz -box_focus_slices all_runs_mprage_ss.nii.gz -cbar ROI_i32 -func_range 32 -pbar_posonly -opacity 9 -blowup 1 -save_ftype JPEG -prefix final_roi_map -montx 6 -monty 3 -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_24.1.01
++ chauffeur ver : 6.7
------------------ start of optionizing ------------------
++ Found input file: anat_final.masters02-baseline+tlrc.HEAD
++ Found input file: final_roi_map.nii.gz
++ Found focus refbox file: all_runs_mprage_ss.nii.gz
++ Using blowup factor: 1
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ./__tmp_chauf_mPHvlOA9Isk
++ Converted 0 to labels
++ Final subbrick indices: -1 -1 -1
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[0%, 98%] -> [0.000000, 1080.322021]
++ 3dcalc: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tmp_chauf_mPHvlOA9Isk/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tmp_chauf_mPHvlOA9Isk/tmp_olay.nii
++ User-entered function range value value (32)
++ Dimensions (xyzt): 161 191 151 1
++ (initial) Slice spacing ordered (x,y,z) is: 8 10 8
++ 3dAutobox: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Auto bbox: x=11..147 y=12..181 z=9..139
++ 3dAutobox: output dataset = ./__tmp_chauf_mPHvlOA9Isk/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=./__tmp_chauf_mPHvlOA9Isk/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_AFN_XJUCcnl_9Zm8k-eFg3UiSQ argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_24.1.01 (Apr 5 2024) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 0.182275
++ Computing output statistics
++ Output dataset /tmp/3dcalc_AFN_XJUCcnl_9Zm8k-eFg3UiSQ+tlrc.BRIK
++ 3050990 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ -4 21 13
++ Will have the ref box central gapord: 7 9 7
------------------- end of optionizing -------------------
-- trying to start Xvfb :858
[1] 19355
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=6x3:7:0:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=6x3:9:0:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=6x3:7:0:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 ROI_i32 -com DO_NOTHING -com SET_SUBBRICKS -1 -1 -1 -com SET_ULAY_RANGE A.all 0.000000 1080.322021 -com SET_FUNC_RANGE 32 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ -4 21 13 -com SAVE_JPEG axialimage ./final_roi_map.axi blowup=1 -com SAVE_JPEG sagittalimage ./final_roi_map.sag blowup=1 -com SAVE_JPEG coronalimage ./final_roi_map.cor blowup=1 -com QUITT ./__tmp_chauf_mPHvlOA9Isk
+/home/sandra/data/Data/automation-test/newcases/masters02.rois/afni_tutorial_rois_sswarper_fixed_ROIs/__tmp_chauf_mPHvlOA9Isk++ Writing one 966x573 image to filter '/home/sandra/abin/cjpeg -quality 95 > ./final_roi_map.axi.jpg'
++ Writing one 1146x453 image to filter '/home/sandra/abin/cjpeg -quality 95 > ./final_roi_map.sag.jpg'
++ Writing one 966x453 image to filter '/home/sandra/abin/cjpeg -quality 95 > ./final_roi_map.cor.jpg'
AFNI QUITTs!
+* Removing temporary image directory './__tmp_chauf_mPHvlOA9Isk'.
[1] + Done Xvfb :858 -screen 0 1024x768x24
++ DONE (good exit)
see: ./final_roi_map*
++ DONE!
sandra@DESKTOP-RJJ49UB:~/data/Data/automation-test/newcases/masters02.rois/afni_tutorial_rois_sswarper_fixed_ROIs$ ./do_02_surfaceize_rois.tcsh
Notice SUMA_AddNodeIndexColumn:
Assuming node indexing
is explicit.
1st row is for node 0
2nd is for node 1, etc.
Notice SUMA_AddNodeIndexColumn:
Assuming node indexing
is explicit.
1st row is for node 0
2nd is for node 1, etc.
Notice SUMA_AddNodeIndexColumn:
Assuming node indexing
is explicit.
1st row is for node 0
2nd is for node 1, etc.
Notice SUMA_AddNodeIndexColumn:
Assuming node indexing
is explicit.
1st row is for node 0
2nd is for node 1, etc.
Notice SUMA_AddNodeIndexColumn:
Assuming node indexing
is explicit.
1st row is for node 0
2nd is for node 1, etc.
Notice SUMA_AddNodeIndexColumn:
Assuming node indexing
is explicit.
1st row is for node 0
2nd is for node 1, etc.
vvvvvvvvvvvvvvvvvvvvvvvvvvvvSurface #1/6(Local Domain Parent), loading ...
++ Notice SUMA_AutoLoad_SO_Dsets (SUMA_Load_Surface_Object.c:1619 @21:53:24):
Auto Loading ./final_roi_map.SURF.left_au.k1.niml.dset onto ./final_roi_map.SURF.left_au.k1.gii
++ Notice SUMA_LoadDsetOntoSO_eng (SUMA_Color.c:10444 @21:53:24):
dset has a mesh parent, Checking relationship
oo Warning SUMA_Contralateral_SO (SUMA_DOmanip.c:2885):
Surface sides are not clearly defined. If this is in error, consider adding
Hemisphere = R (or L or B) in the spec file
to make sure surfaces sides are correctly labeled.
Similar warnings will be muted
vvvvvvvvvvvvvvvvvvvvvvvvvvvvSurface #2/6(Local Domain Parent), loading ...
++ Notice SUMA_AutoLoad_SO_Dsets (SUMA_Load_Surface_Object.c:1619 @21:53:24):
Auto Loading ./final_roi_map.SURF.left_bro.k4.niml.dset onto ./final_roi_map.SURF.left_bro.k4.gii
++ Notice SUMA_LoadDsetOntoSO_eng (SUMA_Color.c:10444 @21:53:24):
dset has a mesh parent, Checking relationship
vvvvvvvvvvvvvvvvvvvvvvvvvvvvSurface #3/6(Local Domain Parent), loading ...
++ Notice SUMA_AutoLoad_SO_Dsets (SUMA_Load_Surface_Object.c:1619 @21:53:24):
Auto Loading ./final_roi_map.SURF.right_au.k2.niml.dset onto ./final_roi_map.SURF.right_au.k2.gii
++ Notice SUMA_LoadDsetOntoSO_eng (SUMA_Color.c:10444 @21:53:24):
dset has a mesh parent, Checking relationship
vvvvvvvvvvvvvvvvvvvvvvvvvvvvSurface #4/6(Local Domain Parent), loading ...
++ Notice SUMA_AutoLoad_SO_Dsets (SUMA_Load_Surface_Object.c:1619 @21:53:24):
Auto Loading ./final_roi_map.SURF.right_bro.k3.niml.dset onto ./final_roi_map.SURF.right_bro.k3.gii
++ Notice SUMA_LoadDsetOntoSO_eng (SUMA_Color.c:10444 @21:53:24):
dset has a mesh parent, Checking relationship
vvvvvvvvvvvvvvvvvvvvvvvvvvvvSurface #5/6(Local Domain Parent), loading ...
++ Notice SUMA_AutoLoad_SO_Dsets (SUMA_Load_Surface_Object.c:1619 @21:53:24):
Auto Loading ./final_roi_map.SURF.right_motor.k5.niml.dset onto ./final_roi_map.SURF.right_motor.k5.gii
++ Notice SUMA_LoadDsetOntoSO_eng (SUMA_Color.c:10444 @21:53:24):
dset has a mesh parent, Checking relationship
vvvvvvvvvvvvvvvvvvvvvvvvvvvvSurface #6/6(Local Domain Parent), loading ...
++ Notice SUMA_AutoLoad_SO_Dsets (SUMA_Load_Surface_Object.c:1619 @21:53:24):
Auto Loading ./final_roi_map.SURF.right_occ.k6.niml.dset onto ./final_roi_map.SURF.right_occ.k6.gii
++ Notice SUMA_LoadDsetOntoSO_eng (SUMA_Color.c:10444 @21:53:24):
dset has a mesh parent, Checking relationship
SUMA_Engine: Starting to listen ...
vvvvvvvvvvvvvvvvvvvvvvvvvvvv
Selected surface final_roi_map.SURF.left_au.k1.gii (Focus_DO_ID # 1).
FaceSet 37, Closest Node 13
Nodes forming closest FaceSet:
25, 13, 14
Coordinates of Nodes forming closest FaceSet:
19.745371, 34.311199, 16.704662
19.695551, 32.293709, 14.874831
22.178751, 34.313274, 13.408155
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
vvvvvvvvvvvvvvvvvvvvvvvvvvvv
Selected surface final_roi_map.SURF.left_bro.k4.gii (Focus_DO_ID # 2).
FaceSet 34, Closest Node 13
Nodes forming closest FaceSet:
13, 25, 10
Coordinates of Nodes forming closest FaceSet:
23.195551, 21.793713, 18.374832
23.245371, 23.811197, 20.204660
21.119585, 22.364252, 17.490644
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
vvvvvvvvvvvvvvvvvvvvvvvvvvvv
Selected surface final_roi_map.SURF.right_bro.k3.gii (Focus_DO_ID # 4).
FaceSet 28, Closest Node 10
Nodes forming closest FaceSet:
13, 10, 9
Coordinates of Nodes forming closest FaceSet:
40.695549, 18.293713, 18.374832
38.619583, 18.864252, 17.490644
40.279606, 17.234304, 17.066328
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
vvvvvvvvvvvvvvvvvvvvvvvvvvvv
Selected surface final_roi_map.SURF.right_motor.k5.gii (Focus_DO_ID # 5).
FaceSet 34, Closest Node 10
Nodes forming closest FaceSet:
13, 25, 10
Coordinates of Nodes forming closest FaceSet:
40.695549, 25.293713, 21.874832
40.745373, 27.311197, 23.704660
38.619583, 25.864252, 20.990644
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
vvvvvvvvvvvvvvvvvvvvvvvvvvvv
Selected surface final_roi_map.SURF.right_occ.k6.gii (Focus_DO_ID # 6).
FaceSet 33, Closest Node 25
Nodes forming closest FaceSet:
25, 26, 10
Coordinates of Nodes forming closest FaceSet:
37.245373, 48.311199, 16.704662
34.848953, 49.659824, 16.506144
35.119583, 46.864254, 13.990644
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
vvvvvvvvvvvvvvvvvvvvvvvvvvvv
Selected surface final_roi_map.SURF.right_au.k2.gii (Focus_DO_ID # 3).
FaceSet 29, Closest Node 13
Nodes forming closest FaceSet:
12, 13, 9
Coordinates of Nodes forming closest FaceSet:
48.565605, 28.822611, 13.133023
47.695549, 28.793711, 14.874831
47.279606, 27.734304, 13.566326