Hello again,
It has been a while since I needed some advice - but my research advisor has been fairly helpful as of late, and only recently has gone silent again for the time being. After much discussion and work, I have been able to (fairly) confidently visualize the data we have.
My only issue now is, the visualization isn't exactly as we were expecting it to be, and I don't know why. It could be due to the resampling method, how I used 3dUndump, or if there's a setting I need to change in the AFNI UI.
Here is the information that is most likely needed:
.txt file format:
This is a slight sample of the data that is being used. The first 3 colums are showing the coordinates (x, y, or z) and the last column is representing a category of stimuli (1, 2, or 3)
-15.449 -12.903 -32.775 1
-46.522 -10.19 -33.231 2
-25.409 -24.693 -28.62 2
44.35 0.53167 -28.234 1
-47.869 31.447 -26.011 1
31.948 38.032 -25.115 1
31.466 41.814 -25.198 2
30.984 45.595 -25.282 2
54.025 15.967 -20.338 2
53.543 19.749 -20.421 3
Resampling File:
The functional mask files from this dataset is taken from my research advisor in our research drive, and is being used to apply the correct resolution to our anatomical template (MNI_caez_N27+tlrc.).
The resampling file contains BRIK/HEADER files for each subject, which are just binary masks, with a 1 everywhere there is cortex in 0 everywhere else. But mainly, they are at the right functional resolution which is the most important part (or so I was told, and am inclined to believe since the visualization is more accurate now). So when I used each subject's mask file as the master file for 3dundump, it will autmoatically then be at the correct resolution.
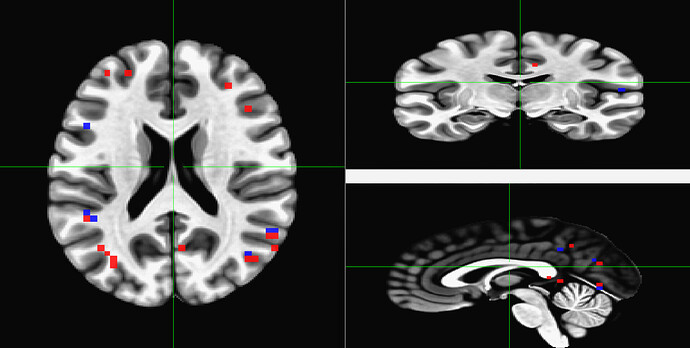
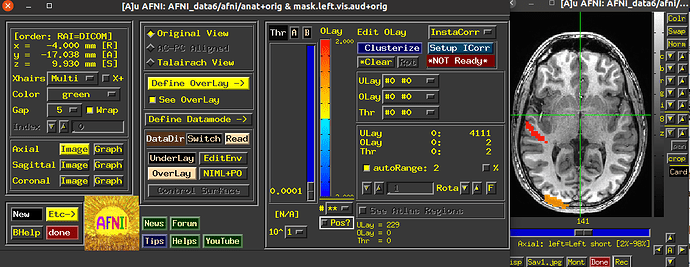
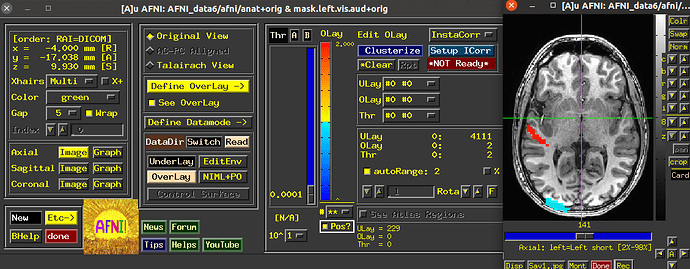
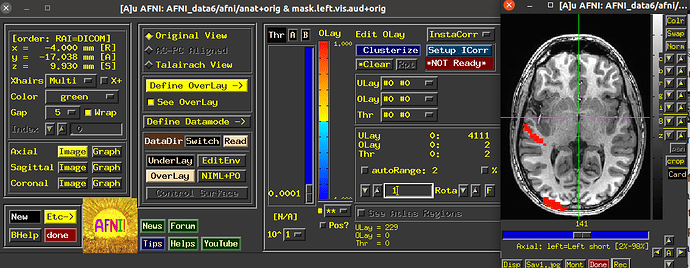
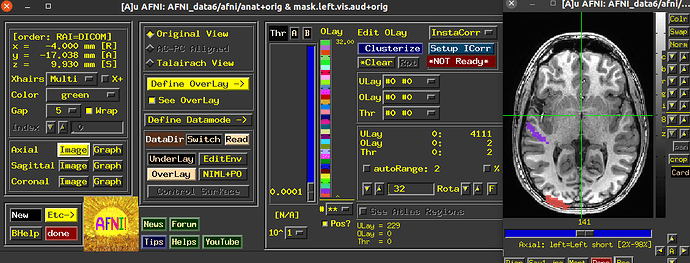
Now after using 3dUndump with these files, and creating a new file that is going to be used in AFNI and then visualized into SUMA with a 2D format so that we can better analyze the patterns, I can only see two of the visual stimuli categories.
Since the range of the data in AFNI is from 0-3, we are expecting each value (1, 2, and 3) to correspond to a color on the visualization; red, blue or green (we are using the RedBlueGreen colorbar) but I am only seeing red and blue, and am unsure why the other stimuli would not be shown.
3dinfo: AFNI version=AFNI_24.2.01 (Jul 16 2024) [64-bit]
Dataset File: ----+tlrc
Identifier Code: ----
Template Space: TLRC
Dataset Type: Anat Bucket (-abuc)
Byte Order: LSB_FIRST [this CPU native = LSB_FIRST]
Storage Mode: BRIK
Storage Space: 245,760 (246 thousand) bytes
Geometry String: "MATRIX(3.75,0,0,-121.4062,0,3.75,0,-108.2812,0,0,4,-40):64,64,30"
Data Axes Tilt: Plumb
Data Axes Orientation:
first (x) = Right-to-Left
second (y) = Anterior-to-Posterior
third (z) = Inferior-to-Superior [-orient RAI]
R-to-L extent: -121.406 [R] -to- 114.844 [L] -step- 3.750 mm [ 64 voxels]
A-to-P extent: -108.281 [A] -to- 127.969 [P] -step- 3.750 mm [ 64 voxels]
I-to-S extent: -40.000 [I] -to- 76.000 [S] -step- 4.000 mm [ 30 voxels]
Number of values stored at each pixel = 1
-- At sub-brick #0 '#0' datum type is short: 0 to 3