AFNI version info (afni -ver): Version AFNI_23.1.05 'Publius Helvius Pertinax'
Hi all,
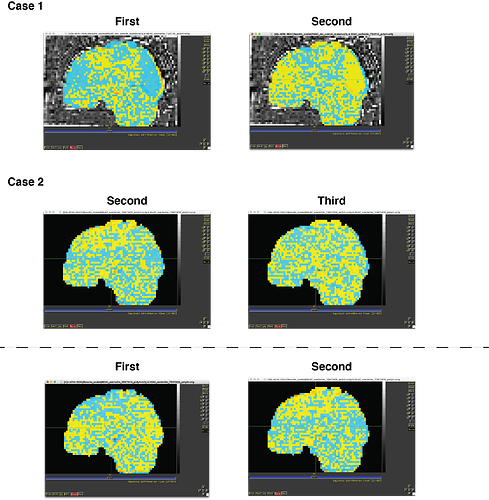
I runned the 3dDeconvolve with TENT function and I found significant low pattern similarity between TR0 and TR1 for certain stimulus.
The stimulus did not showed the significant low pattern similarity is the stimuli that has appeared at the first of the run.
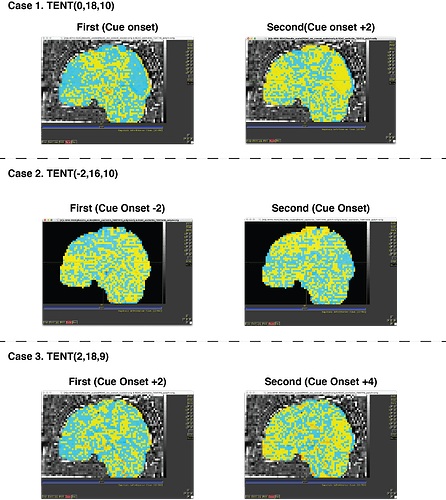
I've tried to change the onset (-2s or +2s, which are -1TR and +1TR) but I could get the same results.
e.g) When cue onset is 10s, the pattern similarity between at the 10s and 12s were too low (near -0.7, -0.8)
When I change the cue onset as -2s, the pattern similarity between the 8s and 10s were too low (near -0.7, -0.8)
When I changed the cue onset as +2s, the pattern similarity between the 12s and 14s were too low (near -0.7, -0.8)
Is there any reseaon that this low pattern similarity could appear? and is this the correct neural activity?
(But I thought this result were too weired, because I could see the reversed activity of entire brain at TR0 and TR1 when I observed the result of the deconvolution by afni)
If this is the limitation of the TENT function, can I deconvolve the neural actvitiy before 1TR from the cue onset, and eliminate the first output to get the neural activity at cue onset?
+) There is one stimuli that is presented at the first of the run, which means onset time is 0s. Is it possible to deconvolve from the 1TR before the onset (which means, using the TENT like TENT(-2,16,10) for the given stimuli time 0). I conducted that, and there was no warning (could be due to there are repeats) but I can not sure for the output of this.
+) I will conduct the time correction with different time range, to make sure there is some values before the 0s onset time at the beginning of the run. (e.g. cutting less bookend TR). Is it better way to get proper output from the deconvolution, if I want to deconvolve 1TR before the onset?
Below part is my script for the deconvolution
#!/bin/tcsh
set analName = REAC
set analDir = $subjectDir/REAC_add1TR
set resultsDir = $analDir/Results_scaled
set stimDir = $analDir/Stim_Concat
set contrastDir = $analDir/Contrasts
echo 1
set inputFile = $resultsDir/${analName}_mc_concat_scaled+orig.HEAD
#set inputFile = $resultsDir/${analName}_mc_concat+orig.HEAD
echo $inputFile
cd $resultsDir
rm ${analName}_eachstim_TENTM10_poly4*
3dDeconvolve -xout -input ${inputFile} \
-xjpeg $resultsDir/matrix \
-mask $maskDir/brainMask+orig \
-polort 4 \
-jobs 32 \
-num_stimts 18 \
-float \
-tout \
-basis_normall 1 \
-stim_times 1 ${stimDir}/${participant}-${analName}-1 'TENT(-2,16,10)' -stim_label 1 1 \
-stim_times 2 ${stimDir}/${participant}-${analName}-2 'TENT(-2,16,10)' -stim_label 2 2 \
-stim_times 3 ${stimDir}/${participant}-${analName}-3 'TENT(-2,16,10)' -stim_label 3 3 \
-stim_times 4 ${stimDir}/${participant}-${analName}-4 'TENT(-2,16,10)' -stim_label 4 4 \
-stim_times 5 ${stimDir}/${participant}-${analName}-5 'TENT(-2,16,10)' -stim_label 5 5 \
-stim_times 6 ${stimDir}/${participant}-${analName}-6 'TENT(-2,16,10)' -stim_label 6 6 \
-stim_times 7 ${stimDir}/${participant}-${analName}-7 'TENT(-2,16,10)' -stim_label 7 7 \
-stim_times 8 ${stimDir}/${participant}-${analName}-8 'TENT(-2,16,10)' -stim_label 8 8 \
-stim_times 9 ${stimDir}/${participant}-${analName}-9 'TENT(-2,16,10)' -stim_label 9 9 \
-stim_times 10 ${stimDir}/${participant}-${analName}-10 'TENT(-2,16,10)' -stim_label 10 10 \
-stim_times 11 ${stimDir}/${participant}-${analName}-11 'TENT(-2,16,10)' -stim_label 11 11 \
-stim_times 12 ${stimDir}/${participant}-${analName}-12 'TENT(-2,16,10)' -stim_label 12 12 \
-stim_file 13 ${resultsDir}/REACMotion'[1]' \
-stim_base 13 \
-stim_file 14 ${resultsDir}/${analName}Motion'[2]' \
-stim_base 14 \
-stim_file 15 ${resultsDir}/${analName}Motion'[3]' \
-stim_base 15 \
-stim_file 16 ${resultsDir}/${analName}Motion'[4]' \
-stim_base 16 \
-stim_file 17 ${resultsDir}/${analName}Motion'[5]' \
-stim_base 17 \
-stim_file 18 ${resultsDir}/${analName}Motion'[6]' \
-stim_base 18 \
-concat $contrastDir/runs_${analName}.1D \
-full_first -fout -tout \
-bucket $resultsDir/${analName}_eachstim_TENTM10_poly4
cd
Best Regards,
Minjae Kwon