AFNI version info (afni -ver): AFNI_24.3.06
Dear AFNI Team,
I ran my first level analysis and for the vast majority of participants I got great Epi to Anat alignment results with sswarper2 and afni_proc.py:
afni_proc.py \
-subj_id ${subj_id}_${sess} \
-copy_anat $anat_dir/anatSS.${subj_id}_${sess}.nii \
-anat_has_skull no \
-anat_follower anat_w_skull anat $anat_dir/anat_T1+orig \
-dsets $epi_dir/epi_nback+orig.HEAD \
-blocks tshift align tlrc volreg mask blur \
scale regress \
-radial_correlate_blocks tcat volreg \
-tcat_remove_first_trs 4 \
-align_unifize_epi local \
-align_opts_aea -cost lpc+ZZ \
-giant_move \
-check_flip \
-tlrc_base MNI152_2009_template_SSW.nii.gz \
-tlrc_NL_warp \
-tlrc_NL_warped_dsets $anat_dir/anatQQ.${subj_id}_${sess}.nii \
$anat_dir/anatQQ.${subj_id}_${sess}.aff12.1D \
$anat_dir/anatQQ.${subj_id}_${sess}_WARP.nii \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-volreg_compute_tsnr yes \
-mask_epi_anat yes \
-blur_size 6.0 \
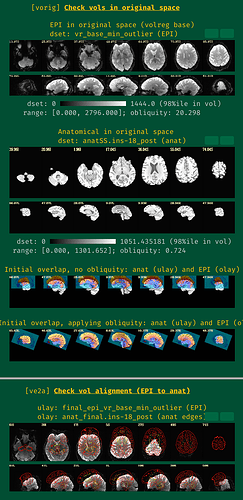
However, for very few subjects the initial alignment between the Epi and anatomical dataset appears to be quite off and the subsequent alignment doesn't seem to improve it - do you have any suggestions how to improve alignment in that case? I already tried several options like -ginormous_move or alternative cost functions.
Here is an example from the QC:
I appreciate any advice!
Best,
Florian