AFNI version info: 24.1.11
Greetings!
I'm following Reynolds' script to complete the PPI analysis. Everything seems to be going well. However, I've noticed that timing_tool.py doesn't seem to be able to properly handle time series files containing two modulators when creating timing partition files:
timing_tool.py -timing $tfile \
-tr $TRup -stim_dur $stim_dur[$sind] \
-run_len $run_lens \
-min_frac 0.3 \
-timing_to_1D $timing_prefix.$sind2.$label \
-per_run_file -show_timing
(Example of my time series file: option times * CR, EV)
![]()
This results in what appears to be a missing coefficent in the final creation of the PPI regression model:
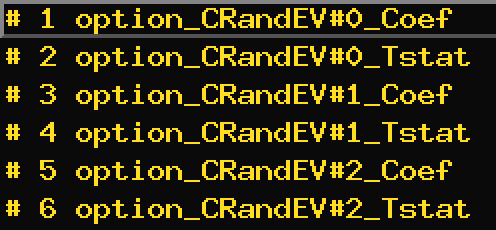

If the regression term labeled 'option_CRandEV' in my original GLM model contains two modulators, then in the regression for PPI, it should also contain the two corresponding PPI coef (is that right?), but here there is only one,
So I can't know the PPI results modulated by option_CR or option_EV alone.
One way I can think of to deal with this is to take files with only 1 amplitude attached to each time when using the stim_times_AM2 option, and to revise the code of 3ddeconvolve :
-stim_times_AM2 1 {m_txt1} 'BLOCK5(1,1)' \
-stim_times_AM2 2 {m_txt2} 'BLOCK5(1,1)' \
-stim_label 1 'optionCRandEV' \
-stim_label 2 'gamleRPE' \
to
-stim_times_AM2 1 {m_txt1} 'BLOCK5(1,1)' \
-stim_times_AM2 2 {m_txt2} 'BLOCK5(1,1)' \
-stim_times_AM2 3 {m_txt3} 'BLOCK5(1,1)' \
-stim_label 1 'optionCR' \
-stim_label 2 'optionEV' \
-stim_label 3 'gamleRPE' \
I don't know if this is the right way to do it. But if anyone has a better suggestion, I would be very grateful!