Hello AFNI experts,
I processed fMRI data using afni_proc.py in the original space. The anatomical image was processed with @SSwarper2, followed by recon-all and @SUMA_Make_Spec_FS.
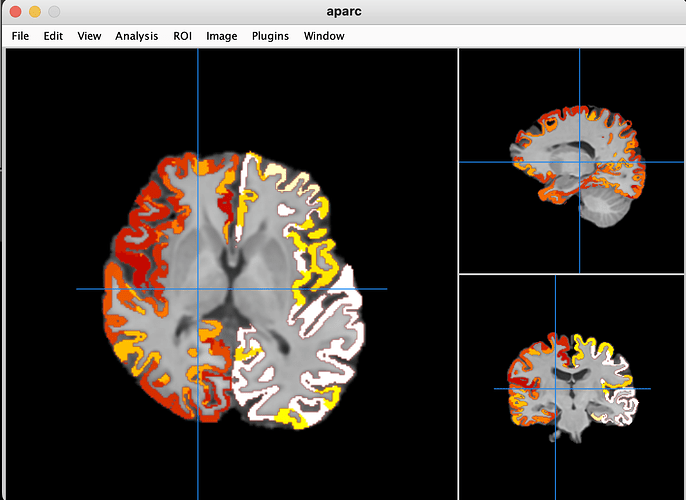
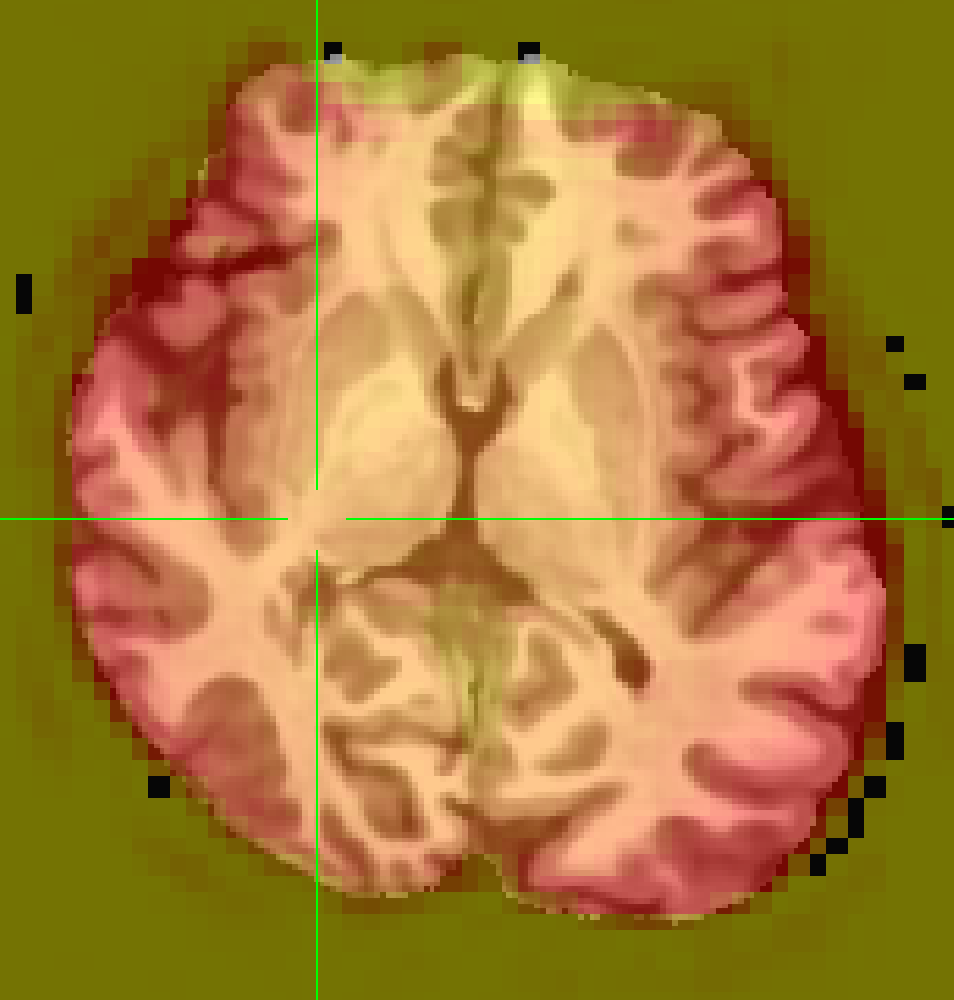
The Freesurfer segmentation file aparc.a2009s+aseg_REN_all.nii.gz appears to align well with anatSS.sub-ep069_anat.nii when viewed in Mango which I assume applies a deoblique transformation automatically.
However, it is not the case when I overlay copy_af_FS_EN_anat on anatSS.sub-ep069 in AFNI
It could also be a false memory, but I recall that copy_af_FS_REN_anat usually aligns well with anatSS when I visulized them in AFNI in another dataset. This is the first time I have seen such a misalignment. Obliquity differs: anatSS has 3.427°, aparc has 0.000°
> 3dinfo -prefix -space -av_space -obliquity -orient -d3 -n4 \
anatSS.sub-ep069_anat.nii \
aparc.a2009s+aseg_REN_all.nii.gz
anatSS.sub-ep069_anat.nii ORIG +orig 3.427 LPI -0.999999 -1.000000 1.000000 176 256 256 1
aparc.a2009s+aseg_REN_all.nii.gz ORIG +orig 0.000 RSP 1.000000-1.000000 -1.000000 256 256 256 1
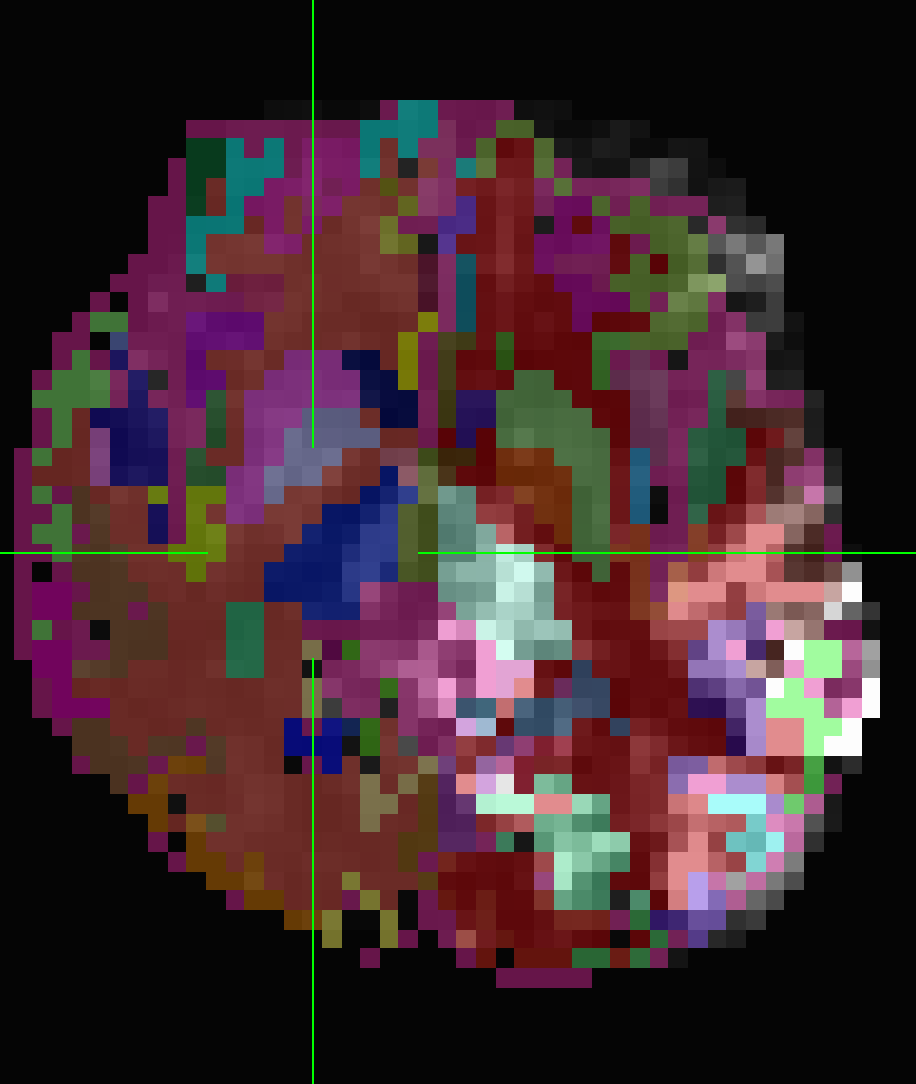
The follow_ROI_FS_REN_epi which was resampled on epi after volreg is also not aligned with anatSS when viewing in AFNI
Same when I overlay follow_ROI_FS_REN_epi on pb02_sub-ep069.r01.volreg
However, I do have the following in the process code which align copy_af_FS_REN_epi+orig to volreg epi
# warp follower dataset copy_af_FS_REN_epi+orig
3dAllineate -source copy_af_FS_REN_epi+orig \
-master pb02.$subj.r01.volreg+orig \
-final NN -1Dparam_apply '1D: 12@0'\' \
-prefix follow_ROI_FS_REN_epi
Then pb02_sub-ep069.r01.volreg is aligned with anatSS...
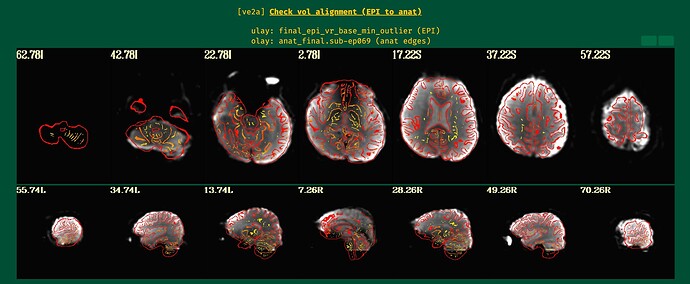
The epi to anat aligment looks fine in the QC
When I overlay follow_ROI_FS_REN_epi on stats.sub-ep069_REML+orig.BRIK.gz they are off, but there is no oblique
> 3dinfo -prefix -space -av_space -obliquity -orient -d3 -n4 \
stats.sub-ep069_REML+orig.BRIK.gz \
follow_ROI_FS_REN_epi+orig.BRIK.gz
stats.sub-ep069_REML ORIG +orig 0.000 LPI -3.500000 -3.500000 3.500000 5073 73 37
follow_ROI_FS_REN_epi ORIG +orig 0.000 LPI -3.500000 -3.500000 3.500000 5073 73 1
Could you please help to understand this behavior of AFNI? Is this just a visualization issue? If everthing is well aligned, is there a recommended way to configure AFNI to show them correctly aligned? If I read BRIK data, follow_ROI_FS_REN_epi+orig.BRIK.gz and stats.subxxx_REML+orig.BRIK.gz using AFNI R script read.AFNI(), are they aligned by voxel 3D index?
Thanks a lot.