Hi-
Well, quality control is a pretty major topic, and involves more than looking at final results. You should check and verify individual steps, which is what the APQC HTML that afni_proc.py creates is meant to help with. We have a few different descriptions of doing QC with AFNI, listed here. Two primary ones might be:
- Reynolds RC, Taylor PA, Glen DR (2023). Quality control practices in FMRI analysis: Philosophy, methods and examples using AFNI. Front. Neurosci. 16:1073800. doi: 10.3389/fnins.2022.1073800
- Taylor PA, Glen DR, Chen G, Cox RW, Hanayik T, Rorden C, Nielson DM, Rajendra JK, Reynolds RC (2024). A Set of FMRI Quality Control Tools in AFNI: Systematic, in-depth and interactive QC with afni_proc.py and more.
Imaging Neuroscience 2: 1–39. doi: 10.1162/imag_a_00246
Those describe things to look at and the mechanics of doing so with the APQC HTML. Here are a few ideas about how to proceed:
- verify FOV and alignment alignment
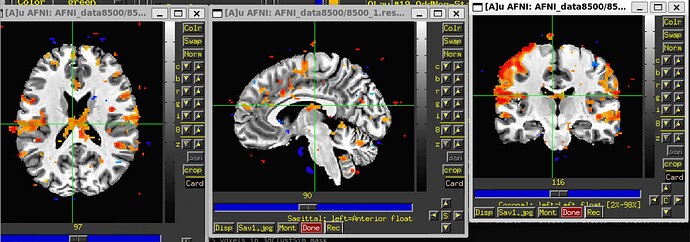
- Look at the EPI-to-anatomical alignment (ve2a) image and make sure you have the coverage and signal coverage where you want it (and see below about TSNR)
- Check the TSNR map and tables to make sure your signal looks strong enough in expected places
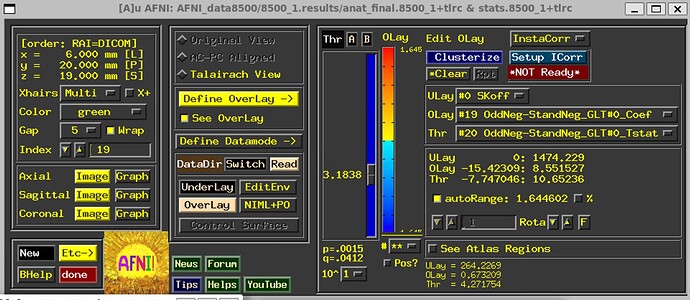
- If you are happy with the alignment steps, for example, then checking the Full F-stat image might help determine whether your created regressors of interest (the timing files and chosen HRF) provide a good fit to observed results in expected locations.
- If those don't, then checking the input timing files and/or HRF might be in order.
- You can also look at the idealized sum of task stimuli, as well as individual ones, to make sure they look reasonable (not duplicated, wrong size, density, etc.).
- Make sure your timing files are appropriately sync'ed and input, and that no missing "chop off this number of initial time points" or anything is missing in your processing
- Checking motion and degrees of freedom counts will be helpful to see if that is affecting your results
- Using InstaCorr to check for potential artifacts might also be helpful (clicking the "IC" button above any of the stats images, or above radcor ones or TSNR ones
As a separate direction, below I have put a copy of your AP command, with aesthetic changes, and I include it here in case it is useful (the options have been options reordered, grouped together by block and vertically spaced).
To compare your inputs with some examples, I ran the command with this opt added, to compare it to a modern task FMRI example based on our Bootcamp class (though Ex. 6b includes nonlinear alignment, which is slower than we use in the Bootcamp example):
-compare_opts 'example 6b'
I didn't see a lot of differences of options, just that the "blur" and "mask" blocks were in opposite order. I don't think that will affect that much in your specific question, because the mask is just calculated not applied during processing. However, your single subject brain mask will likely be larger than the actual brain later, so you might want to change that order (-> that is, do "mask" before "blur" in the block list---nothing else needs to change, because the order of opts doesn't matter, just grouping by block makes things more digestible).
--pt
A copy of the AP command, reordering opts to see what goes together more easily:
afni_proc.py \
-subj_id 8500_1 \
-script proc.8500_1subj \
-scr_overwrite \
-dsets Oddball?_8500.nii \
-copy_anat anatSS.sub-8500.nii \
-anat_has_skull no \
-blocks tshift align tlrc volreg blur mask scale \
regress \
-radial_correlate_blocks tcat volreg regress \
-align_unifize_epi local \
-align_opts_aea -giant_move -cost lpc+ZZ \
-check_flip \
-tlrc_base MNI152_2009_template.nii.gz \
-tlrc_NL_warp \
-tlrc_NL_warped_dsets anatQQ.sub-8500.nii anatQQ.sub-8500.aff12.1D \
anatQQ.sub-8500_WARP.nii \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-volreg_compute_tsnr yes \
-blur_size 4.0 \
-mask_epi_anat yes \
-regress_stim_times Stand_Regress_Correct_Onset_NEU_8500.txt \
Stand_Regress_Correct_Onset_NEG_8500.txt \
Odd_Regress_Correct_Onset_NEU_8500.txt \
Odd_Regress_Correct_Onset_NEG_8500.txt \
Stand_Regress_Incorrect_Onset_NEU_8500.txt \
-regress_stim_labels StandardNeuC StandardNegC OddballNeuC \
OddballNegC StandardNeuX \
-regress_basis 'BLOCK(2,1)' \
-regress_opts_3dD -gltsym 'SYM: OddballNeuC -StandardNeuC' \
-glt_label 1 OddNeu-StandNeu \
-gltsym 'SYM: OddballNegC -StandardNegC' \
-glt_label 2 OddNeg-StandNeg \
-regress_motion_per_run \
-regress_censor_motion 0.3 \
-regress_censor_outliers 0.05 \
-regress_make_ideal_sum sum_ideal.1D \
-regress_est_blur_epits \
-regress_est_blur_errts \
-execute