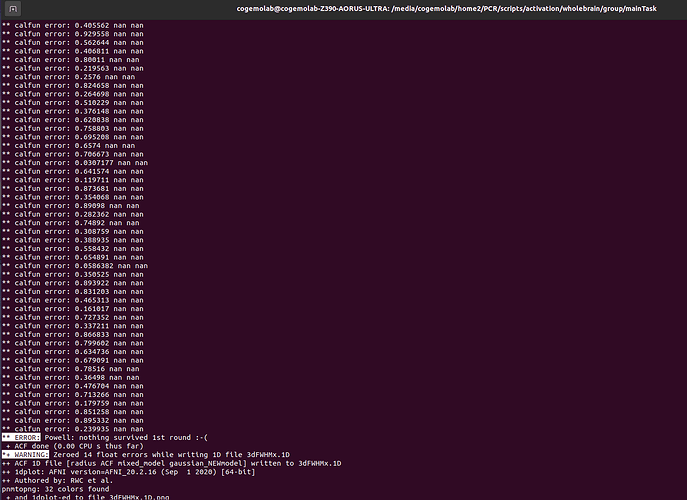
AFNI version info (20.2.16):
Dear AFNI team,
I encountered a challenge while attempting to run 3dClustSim to estimate the probability of false positive clusters. During the calculation of the ACF using 3dFWHMx, I encountered an issue with one participant. Specifically, 3dFWHMx was unable to estimate the 2nd and 3rd ACF values, displaying the error message “Calfun[1]=nan; Calfun[2]=nan” [Attached is a screenshot for your reference].
I am puzzled as to why this error occurred only for one participant. In investigating further, I performed a 3dinfo on the errts.nii file and noticed values in the thousands, despite having done intensity scaling by a factor of 100. Could this indicate an issue with my preprocessing or functional normalization?
If needed, I am more than willing to upload the errts and bucket file for your review and assistance.
Thank you for your time and support.
Best regards,
Sahithyan