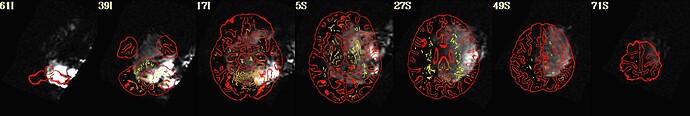
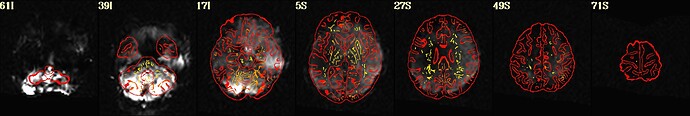
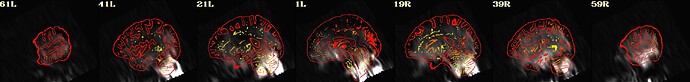
AFNI version info (afni -ver):
Precompiled binary linux_ubuntu_16_64: Jan 31 2024 (Version AFNI_24.0.04 'Caracalla')
Dear AFNI experts,
As hesitant as I am to post this request for help publicly, I'm basically at my wit's end with trying to align a noisy EPI scan from a stroke survivor with the corresponding anatomy. Here's what I found works well enough just using align_epi_anat.py:
# EPI to FLAIR alignmet seems to basically work.
# But first need to align T1 and FLAIR scans.
# FLAIR to Anat:
align_epi_anat.py -dset1to2 -dset1 flair_CM.nii.gz -dset2 anat_CM.nii.gz -cost lpc+ -big_move -overwrite
# EPI to FLAIR to Anat
align_epi_anat.py -anat flair_CM_al+orig \
-epi2anat -epi run2_CM.nii.gz \
-epi_base median \
-giant_move \
-cost lpa+ZZ \
-epi_strip None \
-resample off \
-overwrite
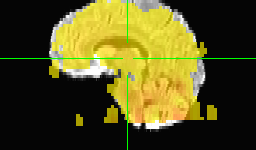
However, ultimately I want to run afni_proc.py to pre-process the data for eventual functional connectivity analysis, per example 11 in the afni_proc.py help. For that, my understanding is that it's best to do all the pre-processing in one shot. But I can't figure out how to do the equivalent of the align_epi_anat.py commands above in afni_proc.py. When I try this, the epi-anat alignment doesn't look so good:
afni_proc.py \
-subj_id $subj \
-copy_anat anat_warped/anatSS.${subj}.nii \
-blocks despike tshift align tlrc volreg blur mask \
scale regress \
-anat_has_skull no \
-anat_follower anat_w_skull anat $subj/SUMA/${subj}_SurfVol.nii \
-anat_follower_ROI aaseg anat $subj/SUMA/aparc.a2009s+aseg.nii.gz \
-anat_follower_ROI aeseg epi $subj/SUMA/aparc.a2009s+aseg.nii.gz \
-anat_follower_ROI FSvent epi $subj/SUMA/fs_ap_latvent.nii.gz \
-anat_follower_ROI FSWe epi $subj/SUMA/fs_ap_wm.nii.gz \
-anat_follower_erode FSvent FSWe \
-dsets run1_CM.nii.gz run2_CM.nii.gz run3_CM.nii.gz run4_CM.nii.gz \
-tcat_remove_first_trs 2 \
-align_unifize_epi yes \
-align_opts_aea -anat flair_CM_al+orig \
-epi2anat \
-epi_strip None \
-epi_base median \
-cost lpa+ZZ \
-giant_move \
-resample off \
-check_flip \
-tlrc_base /usr/local/afni/bin/MNI152_2009_template_SSW.nii.gz \
-tlrc_NL_warp \
-tlrc_NL_warped_dsets anat_warped/anatQQ.$subj.nii anat_warped/anatQQ.$subj.aff12.1D anat_warped/anatQQ.${subj}_WARP.nii \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-blur_size 4 \
-mask_epi_anat yes \
-regress_motion_per_run \
-regress_ROI_PC FSvent 3 \
-regress_ROI_PC_per_run FSvent \
-regress_make_corr_vols aeseg FSvent \
-regress_anaticor_fast \
-regress_anaticor_label FSWe \
-regress_censor_motion 1.0 \
-regress_censor_outliers 0.3 \
-regress_apply_mot_types demean deriv \
-regress_est_blur_epits \
-regress_est_blur_errts \
-scr_overwrite \
-html_review_style pythonic \
-execute
Paul, I plan to follow your advice from another thread and remove the "blur" references later. But for now I'm just trying to get the basics to work.
-Will