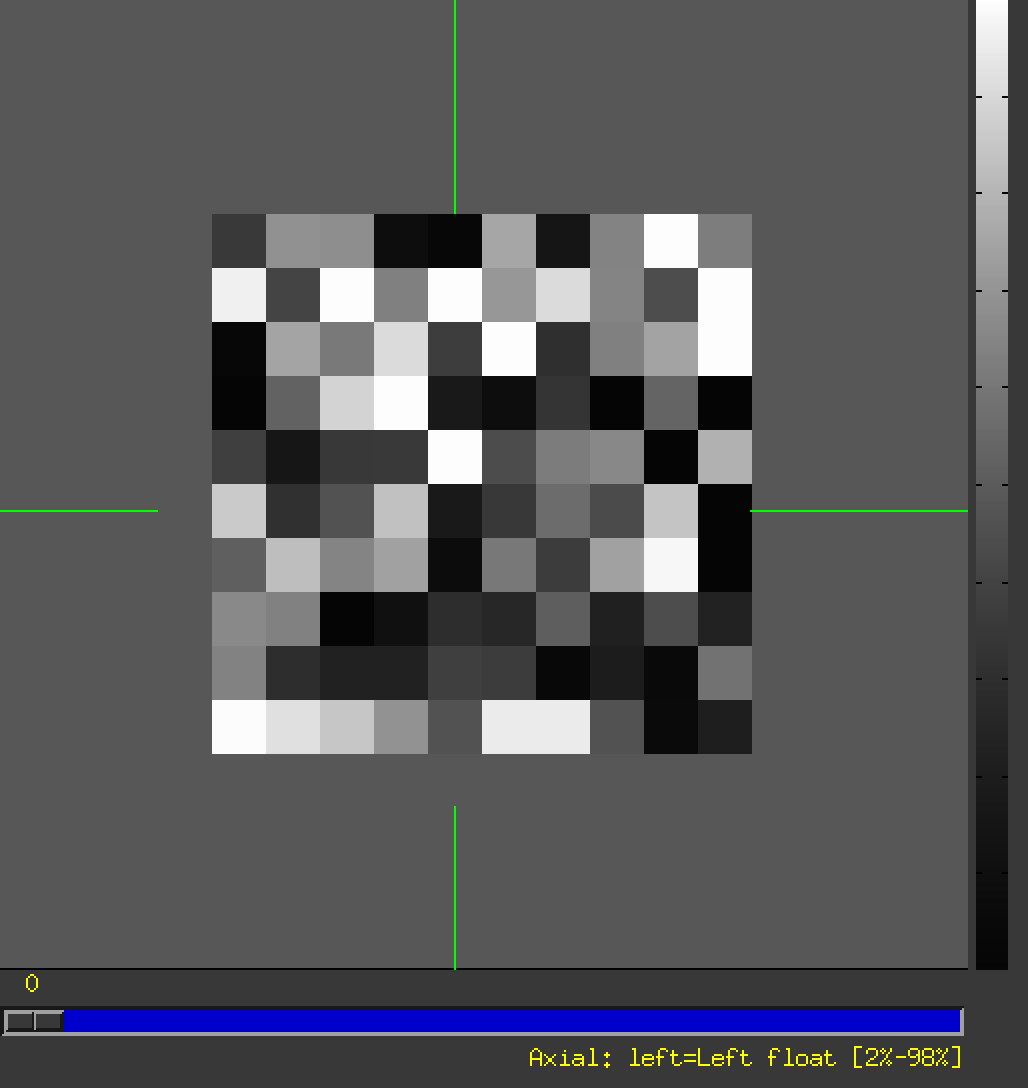Dear Afni experts,
Is is possible to provide 3dAllineate with some custom starting transformation? The idea is to avoid applying these starting transformations (one per frame) before feeding the resulting volume timeseries to 3dAllineate--avoiding extra interpolation and read/write.
Thanks and
Have a great day!
Sébastien
Hi, Sébastien-
Can I ask what the context of this is? Like, is this just in some script you are writing, or within afni_proc.py usage? That might affect some implementation ideas.
Without knowing any other info, and assuming this is in a standalone script doing alignment, I would just apply your initial data, and then take the result as the input/source for 3dAllineate. When you are done, you can concatenate your pre-existing transform and the estimated one, to apply in the future to the initial data (and hence avoid extra interpolation).
I would note that align_epi_anat.py, which wraps around 3dAllineate, has the -pre_matrix .. option which does what you are asking. So, if you are doing this within the context of afni_proc.py, that will be useful because AP uses align_epi_anat.py for the affine EPI-anatomical alignment.
--pt
Thanks for the quick answer Paul!
Yes I am writing my own scripts to process weird fMRI data--single-slice high-resolution FLASH for vessel fMRI where static tissue is highly suppressed and vessels very bright. I'm wrapping 3dAllineate--not 3dvolreg just to have more options---to perform motion correction in successive steps, and indeed I concatenate the transforms for a final one-step interpolation of the original data.
The ideal algorithm for these data is not yet clear so I'm playing around a lot. I noticed 3dAllineate's coarse and fine steps and I supposed that passing the results of the first step to the second is done in some smart way that avoids reading the data again. So I wondered whether I was missing a perhaps secret option to pass the transform from my own first pass directly to 3dAllineate's second pass, with the goal of mostly just speeding things up.
Now that might be a very small win since as I wrote this I realize that only the need to write the data out of my first step can be removed, not the need to read it again for the second step... and I doubt interpolation affects the estimation of motion in the second steps at all.
Conclusion, don't bother if there is not already a secret way into 3dAllineate's fine step. I'm perfectly fine with chaining algorithmic steps outside 3dAllineate.
Thanks again for the info and
Have a great day!
Sébastien
OK, that extra description helps. We have applied 3dAllineate to work on slicewise data before, and actually I think it was venous stuff specifically... I'm digging around to see if I can find the script, but I am not successful so far.
Things to note about this: Probably the data goes to the edge of your planar dset, rather than being surrounded by empty space like most human brain dsets (except for that pesky neck that we aren't allowed to do anything about). During motion correction, you want to have the nonzero data free move around a bit (and hence be motion corrected). The danger when there is data to the edge of the boundary is that things aren't really allowed to move freely. So you can zeropad to create space around the data, to let the nonzero part move freely. But, there is a risk then that "motion correction" will just align the squares with data to the squares with data---inherently doing nothing. So, we add a weight mask with feathered edges to help the alignment be driven by internal structures of the data and not just boundaries.
Outline of steps, as I recall:
- use 3dZeropad to put some zero padding around the planar dsets' edges (say, 4 voxels deep on each edge), and let's call this
rslice_INFL.nii. An example to do this, if your planar data is in the RL and AP planes, is: 3dZeropad -R 4 -L 4 -A 4 -P 4 -prefix rslice_INFL.nii rslice.nii
- to make the weighted+feathered mask dataset, do something like this with one of hte inflated datasets:
# make an unscaled, float-valued mask where there is nonzero data in 'a'
3dcalc -a rslice_INFL.nii"[0]" -expr 'bool(a)' -prefix rslice_MASK.nii -datum float -nscale
# smooth out the edges of hte weight mask.
3dmerge -1blur_sigma 2.0 -prefix rslice_WEIGHT.nii rslice_MASK.nii
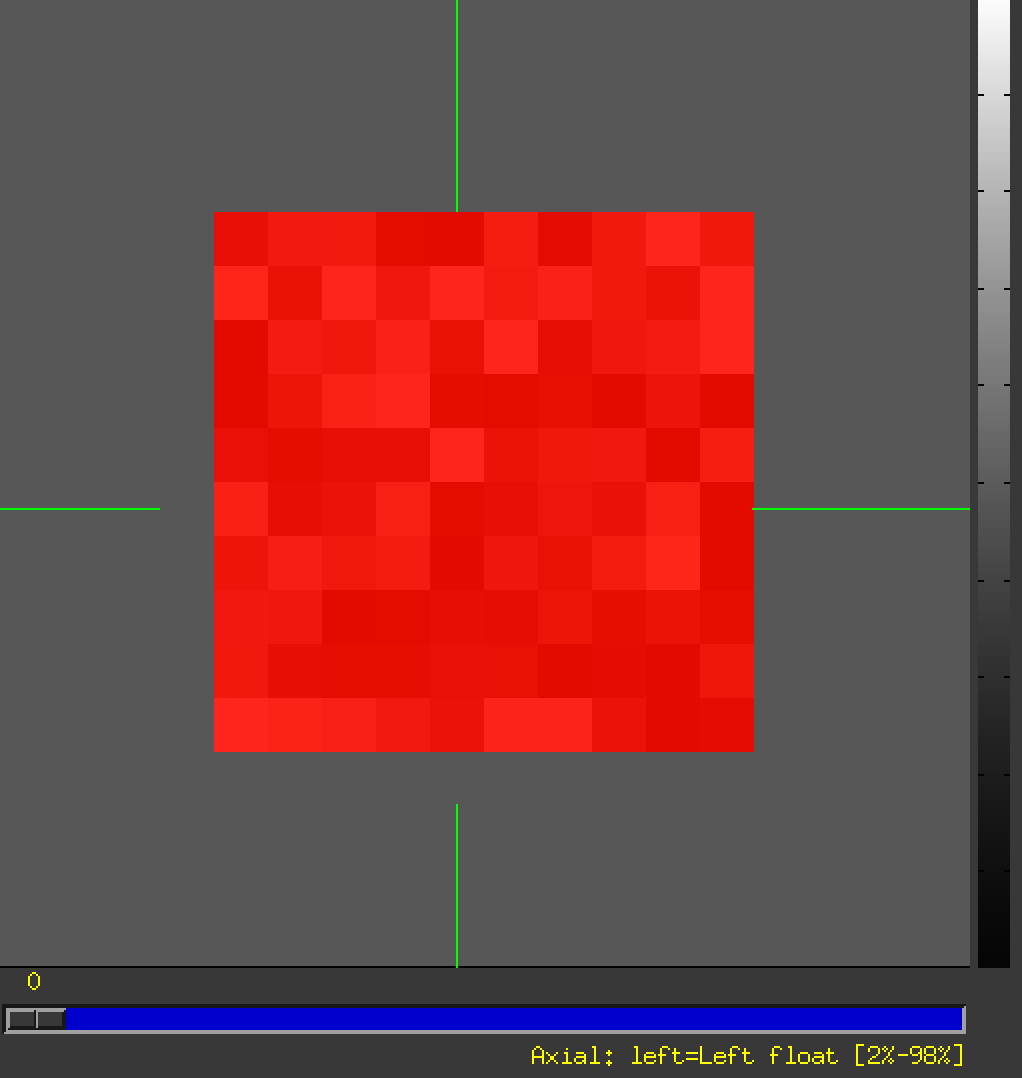
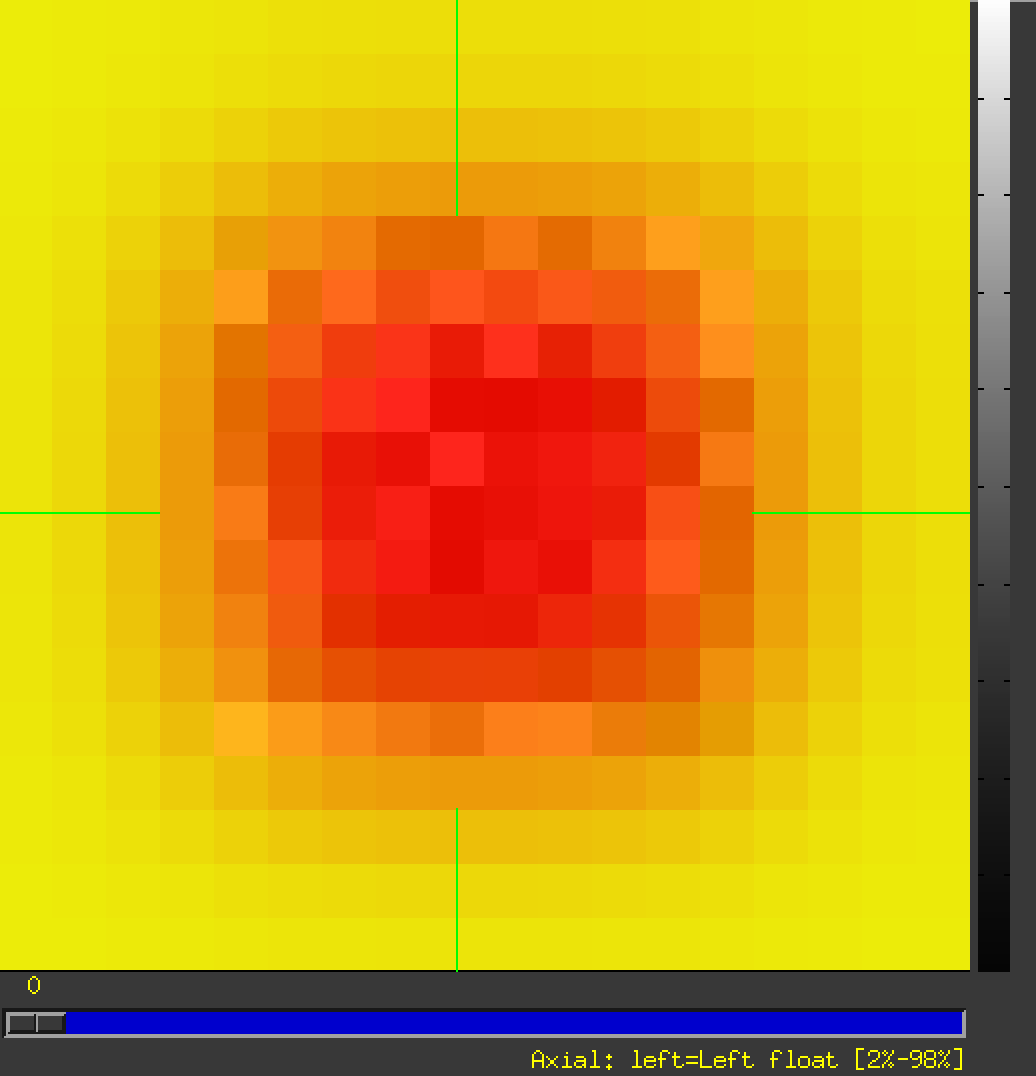
My data successfully look like:
- the rslice_INFL.nii (as underlay):
- ... and overlay the rslice_MASK.nii:
- ... and change the overlay to the rslice_WEIGHT.nii:
Now, you can use 3dAllineate with that -weight rslice_WEIGHT.nii as a weightmask, and use 'lpa' cost function for the alignment. You can decide if restricting alignment to rigid-body alignment makes sense---that will simplify the motion correction; to do so, add -warp shift_rotate.
Please let me know if that makes sense/helps. I'm happy to try this with a bit of the data, too, in case I have missed something important.
--pt
... and I should note in the above that in my mini-test dataset, my voxel size was 1x1 mm**2, and I just doubled that for the blur value sigma in the 3dmerge command. There is probably some flexibility in that choice (as with the number of layers in the 3dZeropad padding), but a factor of 2 is pretty reasonable to give as a first try, I think.
--pt