AFNI version info (afni -ver):
22
I have preprocessed my data with fMRIprep added into AFNI for smoothing and scaling (mean BOLD signal is 100). ROI are a social brain mask from neurosynth.
I am interested in two main contrasts (full second-level code below)...
- Brain differences during exclusion (exclusion block > equal play) between groups
- interaction between groups and age during exclusion (exclusion block > equal play)
My model includes also scanner site (2 locations)
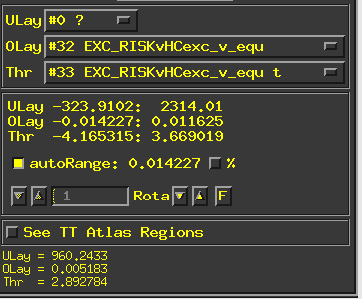
- Are these the correct contrasts to identify brain differences between groups during exclusion vs. equal play (ignore the numbers as they are slightly different to the code below). My understanding is that the first "Olay" box should be the coefficient and the second "Thr" box should be the t-test? This is very different to SPM, so want to make sure I am doing this correctly?
- this is the correct contrast for the age x group comparison?
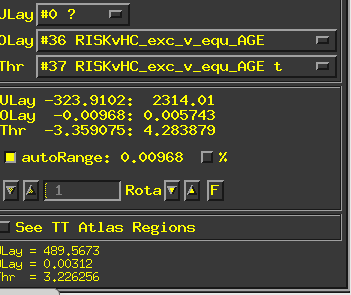
- If I also ran a group-specific contrast of to better understand the interaction findings such as ...
group: 1*HC condition : 1*exclude -1*equal age :'
Would an increased brain response be associated with increased age for this group? or the other way around?
- I need to output the effect size for these contrasts. I specified -GES but don't understand how to output effect size for these contrasts? Could someone explain for a beginner where these values live in AFNI and how to output?
Full code below.
3dMVM -prefix mask_Oct_14_2023 \
-bsVars "grp*age*site" \
-mask '/work/06953/jes6785/atlas_masks/resample_bin_combine_soc_reward_2.nii.gz' \
-wsVars "condition" \
-qVars "age" \
-qVarsCenters '18.4' \
-GES \
-num_glt 2 \
-gltLabel 1 EXC_RISKvHC exc_v_equ -gltCode 1 'grp : 1*RISK -1*HC condition : 1*exclude -1*equal' \
-gltLabel 2 RISKvHC_exc_v_equ_AGE -gltCode 2 'grp : 1*RISK -1*HC condition : 1*exclude -1*equal age :' \
-dataTable \
Subj grp age site condition InputFile \
a002 HC 18.4 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a002b+tlrc[4] \
a004 HC 14.6 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a004b+tlrc[4] \
a042 HC 19 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a042b+tlrc[4] \
a116 HC 20.5 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a116b+tlrc[4] \
a130 HC 19.8 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a130b+tlrc[4] \
a134 HC 18.4 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a134b+tlrc[4] \
a146 HC 15.8 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a146b+tlrc[4] \
b010 HC 18.3 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b010b+tlrc[4] \
b100 HC 20.2 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b100b+tlrc[4] \
b132 HC 15.2 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b132b+tlrc[4] \
b140 HC 20.3 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b140b+tlrc[4] \
a030 RISK 16.4 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a030b+tlrc[4] \
a032 RISK 14.9 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a032b+tlrc[4] \
a034 RISK 14.2 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a034b+tlrc[4] \
a052 RISK 15.8 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a052b+tlrc[4] \
a058 RISK 17.7 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a058b+tlrc[4] \
a060 RISK 16 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a060b+tlrc[4] \
b014 RISK 14.4 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b014b+tlrc[4] \
b022 RISK 15.4 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b022b+tlrc[4] \
b048 RISK 18.2 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b048b+tlrc[4] \
b054 RISK 17.8 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b054b+tlrc[4] \
b074 RISK 19.5 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b074b+tlrc[4] \
b144 RISK 19.6 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b144b+tlrc[4] \
b148 RISK 14.7 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b148b+tlrc[4] \
b150 RISK 17.5 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b150b+tlrc[4] \
b154 RISK 17.2 UC equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b154b+tlrc[4] \
a001 HC 21.1 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a001+tlrc[4] \
a017 HC 21.5 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a017+tlrc[4] \
a025 HC 18.9 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a025+tlrc[4] \
a045 HC 18.5 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a045+tlrc[4] \
a101 HC 20.5 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a101+tlrc[4] \
a105 HC 20.9 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a105+tlrc[4] \
a119 HC 19.8 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a119+tlrc[4] \
a129 HC 20.9 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a129+tlrc[4] \
a131 HC 20.7 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a131+tlrc[4] \
a145 HC 21.3 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a145+tlrc[4] \
b007 HC 21.9 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b007+tlrc[4] \
b009 HC 18.8 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b009+tlrc[4] \
b117 HC 18.6 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b117+tlrc[4] \
b149 HC 14 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b149+tlrc[4] \
b161 HC 19.7 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b161+tlrc[4] \
b163 HC 20.7 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b163+tlrc[4] \
b165 HC 20.5 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b165+tlrc[4] \
b167 HC 21.1 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b167+tlrc[4] \
b173 HC 21.7 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b173+tlrc[4] \
a041 RISK 20 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a041+tlrc[4] \
a073 RISK 19.7 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a073+tlrc[4] \
a087 RISK 19.2 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a087+tlrc[4] \
a135 RISK 17.3 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a135+tlrc[4] \
a141 RISK 17.7 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a141+tlrc[4] \
a147 RISK 18.9 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a147+tlrc[4] \
a153 RISK 19 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a153+tlrc[4] \
b029 RISK 18.8 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b029+tlrc[4] \
b065 RISK 17.2 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b065+tlrc[4] \
b071 RISK 20.3 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b071+tlrc[4] \
b075 RISK 19.9 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b075+tlrc[4] \
b083 RISK 14.3 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b083+tlrc[4] \
b095 RISK 18.6 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b095+tlrc[4] \
b103 RISK 19.7 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b103+tlrc[4] \
b123 RISK 20.5 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b123+tlrc[4] \
b143 RISK 14.1 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b143+tlrc[4] \
b155 RISK 17.3 UT equal /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b155+tlrc[4] \
a002 HC 18.4 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a002b+tlrc[10] \
a004 HC 14.6 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a004b+tlrc[10] \
a042 HC 19 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a042b+tlrc[10] \
a116 HC 20.5 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a116b+tlrc[10] \
a130 HC 19.8 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a130b+tlrc[10] \
a134 HC 18.4 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a134b+tlrc[10] \
a146 HC 15.8 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a146b+tlrc[10] \
b010 HC 18.3 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b010b+tlrc[10] \
b100 HC 20.2 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b100b+tlrc[10] \
b132 HC 15.2 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b132b+tlrc[10] \
b140 HC 20.3 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b140b+tlrc[10] \
a030 RISK 16.4 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a030b+tlrc[10] \
a032 RISK 14.9 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a032b+tlrc[10] \
a034 RISK 14.2 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a034b+tlrc[10] \
a052 RISK 15.8 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a052b+tlrc[10] \
a058 RISK 17.7 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a058b+tlrc[10] \
a060 RISK 16 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a060b+tlrc[10] \
b014 RISK 14.4 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b014b+tlrc[10] \
b022 RISK 15.4 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b022b+tlrc[10] \
b048 RISK 18.2 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b048b+tlrc[10] \
b054 RISK 17.8 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b054b+tlrc[10] \
b074 RISK 19.5 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b074b+tlrc[10] \
b144 RISK 19.6 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b144b+tlrc[10] \
b148 RISK 14.7 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b148b+tlrc[10] \
b150 RISK 17.5 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b150b+tlrc[10] \
b154 RISK 17.2 UC exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b154b+tlrc[10] \
a001 HC 21.1 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a001+tlrc[10] \
a017 HC 21.5 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a017+tlrc[10] \
a025 HC 18.9 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a025+tlrc[10] \
a045 HC 18.5 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a045+tlrc[10] \
a101 HC 20.5 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a101+tlrc[10] \
a105 HC 20.9 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a105+tlrc[10] \
a119 HC 19.8 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a119+tlrc[10] \
a129 HC 20.9 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a129+tlrc[10] \
a131 HC 20.7 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a131+tlrc[10] \
a145 HC 21.3 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a145+tlrc[10] \
b007 HC 21.9 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b007+tlrc[10] \
b009 HC 18.8 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b009+tlrc[10] \
b117 HC 18.6 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b117+tlrc[10] \
b149 HC 14 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b149+tlrc[10] \
b161 HC 19.7 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b161+tlrc[10] \
b163 HC 20.7 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b163+tlrc[10] \
b165 HC 20.5 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b165+tlrc[10] \
b167 HC 21.1 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b167+tlrc[10] \
b173 HC 21.7 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b173+tlrc[10] \
a041 RISK 20 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a041+tlrc[10] \
a073 RISK 19.7 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a073+tlrc[10] \
a087 RISK 19.2 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a087+tlrc[10] \
a135 RISK 17.3 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a135+tlrc[10] \
a141 RISK 17.7 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a141+tlrc[10] \
a147 RISK 18.9 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a147+tlrc[10] \
a153 RISK 19 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.a153+tlrc[10] \
b029 RISK 18.8 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b029+tlrc[10] \
b065 RISK 17.2 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b065+tlrc[10] \
b071 RISK 20.3 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b071+tlrc[10] \
b075 RISK 19.9 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b075+tlrc[10] \
b083 RISK 14.3 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b083+tlrc[10] \
b095 RISK 18.6 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b095+tlrc[10] \
b103 RISK 19.7 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b103+tlrc[10] \
b123 RISK 20.5 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b123+tlrc[10] \
b143 RISK 14.1 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b143+tlrc[10] \
b155 RISK 17.3 UT exclude /scratch/06953/jes6785/SOBP/CYB_SOBP_SEPT_2023/level_1_output/stats.b155+tlrc[10]