Hi, heatherb-
Thanks, and that is a good suggestion for people using @SSwarper, which is generally our currently recommended way for people to align a subject anatomical to a template, such as the MNI. However, @SSwarper includes both linear affine and nonlinear warp components, and I only see linear affine pieces included in your inverse transformation. Also, the initial question was about warping and ROI, specifically, which has the particular constraint of wanting to use an “NN” interpolant when applying the transformation (to preserve integer values in the source dset); in your example, that interpolant is not listed, so I don’t think the integer-valued nature of the input atlas dset would be preserved.
Here is a script that I would use to map ROIs to subj anat space when using @SSwarper, with the above considerations applied. It would run in the Bootcamp Dataset directory: ~/AFNI_data6/FT_analysis/Qwarp/anat_warped/, assuming that you have also added the MNI_Glasser_HCP_v1.0_LPI_2009c.nii.gz in that directory
(which could be downloaded here: https://afni.nimh.nih.gov/pub/sscc/staff/ptaylor/MNI_Glasser_HCP_v1.0_LPI_2009c.nii.gz); or any atlas on the MNI*SSW.nii.gz grid will work there.
#!/bin/tcsh
# -------------------------- input info -----------------------------
# subject anatomy, native (subject) space
set dset_anat = anatUA.FT.nii
# MNI "Glasser" atlas (on grid of MNI*SSW*nii.gz), which might need to
# be downloaded, Dear Reader. NB: any atlas on MNI*SSW*nii.gz grid
# will work here
set dset_atl = MNI_Glasser_HCP_v1.0_LPI_2009c.nii.gz
# SSW transform pieces: affine and NL parts
set ssw_aff = anatQQ.FT.aff12.1D
set ssw_nl = anatQQ.FT_WARP.nii
# ------------------------ outputs to be made ---------------------------
# prefix of output warp: could be made using a ${subj} variable
set ssw_full_inv = anatQQ.FT_full_INVWARP.nii.gz
# ... and to be created, in subj anat space
set dset_atl_subj = mni_glass_in_FT.nii.gz
# ----------------------------------------------------------------------
# create full inv warp: (aff + nl)^{-1}
3dNwarpCat \
-iwarp \
-warp1 "${ssw_nl}" \
-warp2 "${ssw_aff}" \
-prefix "${ssw_full_inv}"
# apply full inv warp, with NN interpolant to preserve ints
3dNwarpApply \
-prefix "${dset_atl_subj}" \
-nwarp "${ssw_full_inv}" \
-ainterp NN \
-source "${dset_atl}" \
-master "${dset_anat}"
# bonus nicety: attach any labletables/atlas points, as well as have
# it open a nice "ROI-like" colorbar in GUI when overlaid
3drefit -copytables "${dset_atl}" "${dset_atl_subj}"
3drefit -cmap INT_CMAP "${dset_atl_subj}"
All the inputs and outputs are listed at the top of the script. I called the script “do_inv_ssw_roi.tcsh”, and it can be run in the Bootcamp data directory like:
tcsh do_inv_ssw_roi.tcsh
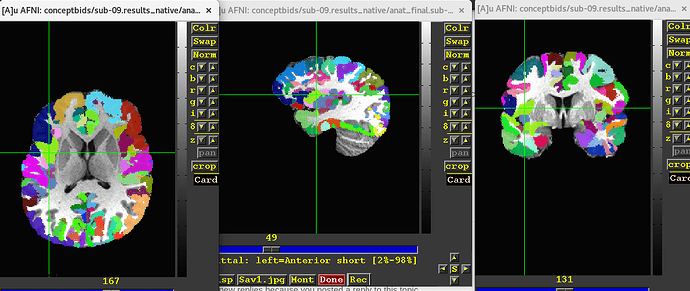
It produced the ROIs as desired, see the attached image, which shows even the labels are attached again successfully.
Yet another consideration might be applying “modal smoothing” to the ROIs after warping like this: basically, because we are mapping one discrete grid to another, some odd edge things can happen, and modal smoothing might be nice to apply. But perhaps seeing how the first steps above seem would be a good place to start.
–pt