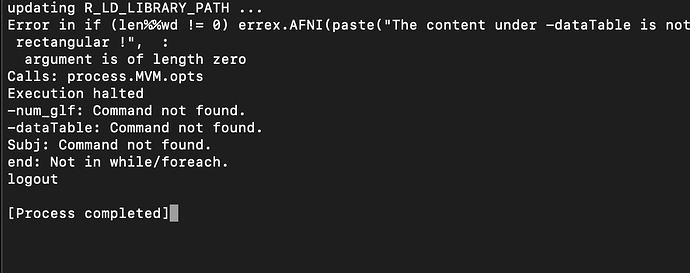
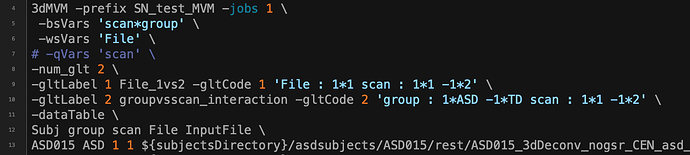
Dimensions:
rows: 75 | columns: 5
Data summary:
Variable Detected_Type Details
Subj Subjects Num Subjects=75
group Categorical Counts: ASD=37 | TD=38
scan Quantitative Min=1 | Max=2 | ? categorical ? Num levels=2
File Quantitative Min=1 | Max=1 | ? categorical ? Num levels=1
InputFile Data Number of InputFiles=75
++ Good: All InputFiles exist.
++ Good: All InputFiles have exactly 1 volume.
++ Good: All InputFiles are on the same grid.
Loading required package: lme4
Loading required package: Matrix
Welcome to afex. For support visit: http://afex.singmann.science/
- Functions for ANOVAs: aov_car(), aov_ez(), and aov_4()
- Methods for calculating p-values with mixed(): 'S', 'KR', 'LRT', and 'PB'
- 'afex_aov' and 'mixed' objects can be passed to emmeans() for follow-up tests
- Get and set global package options with: afex_options()
- Set sum-to-zero contrasts globally: set_sum_contrasts()
- For example analyses see: browseVignettes("afex")
Attaching package: ‘afex’
The following object is masked from ‘package:lme4’:
lmer
Loading required package: car
Loading required package: carData
++++++++++++++++++++++++++++++++++++++++++++++++++++
***** Summary information of data structure *****
75 subjects : 42003 42004 42007 42008 42009 42010 42013 42015 42016 42019 42022 42023 42027 42029 42030 42031 42033 42035 42036 42038 42039 42040 42041 42042 ASD015 ASD022 ASD029 ASD033 ASD034 ASD036 ASD037 ASD042 ASD046 ASD055 ASD056 ASD060 ASD067 ASD079 ASD148E ASD167C ASD168C ASD171C ASD181C ASD184C ASD192C ASD194C ASD200C ASD204 ASD205B ASD207B ASD300 ASD301 ASD302 ASD303 ASD304 ASD305 ASD306 ASD307 ASD308 ASD4004 ASD4005 TD001 TD004 TD012 TD022 TD029 TD031 TD033 TD035 TD043 TD045 TD051 TD053 TD106E TD163C
75 response values
2 levels for factor group : ASD TD
2 levels for factor scan : 1 2
1 levels for factor File : 1
2 post hoc tests
Contingency tables of subject distributions among the categorical variables:
, , File = 1
group
scan ASD TD
1 14 14
2 23 24
Tabulation of subjects against each of the categorical variables:
lop$nSubj vs group:
ASD TD
42003 0 1
42004 0 1
42007 0 1
42008 0 1
42009 0 1
42010 0 1
42013 0 1
42015 0 1
42016 0 1
42019 0 1
42022 0 1
42023 0 1
42027 0 1
42029 0 1
42030 0 1
42031 0 1
42033 0 1
42035 0 1
42036 0 1
42038 0 1
42039 0 1
42040 0 1
42041 0 1
42042 0 1
ASD015 1 0
ASD022 1 0
ASD029 1 0
ASD033 1 0
ASD034 1 0
ASD036 1 0
ASD037 1 0
ASD042 1 0
ASD046 1 0
ASD055 1 0
ASD056 1 0
ASD060 1 0
ASD067 1 0
ASD079 1 0
ASD148E 1 0
ASD167C 1 0
ASD168C 1 0
ASD171C 1 0
ASD181C 1 0
ASD184C 1 0
ASD192C 1 0
ASD194C 1 0
ASD200C 1 0
ASD204 1 0
ASD205B 1 0
ASD207B 1 0
ASD300 1 0
ASD301 1 0
ASD302 1 0
ASD303 1 0
ASD304 1 0
ASD305 1 0
ASD306 1 0
ASD307 1 0
ASD308 1 0
ASD4004 1 0
ASD4005 1 0
TD001 0 1
TD004 0 1
TD012 0 1
TD022 0 1
TD029 0 1
TD031 0 1
TD033 0 1
TD035 0 1
TD043 0 1
TD045 0 1
TD051 0 1
TD053 0 1
TD106E 0 1
TD163C 0 1
lop$nSubj vs scan:
1 2
42003 0 1
42004 0 1
42007 0 1
42008 0 1
42009 0 1
42010 0 1
42013 0 1
42015 0 1
42016 0 1
42019 0 1
42022 0 1
42023 0 1
42027 0 1
42029 0 1
42030 0 1
42031 0 1
42033 0 1
42035 0 1
42036 0 1
42038 0 1
42039 0 1
42040 0 1
42041 0 1
42042 0 1
ASD015 1 0
ASD022 1 0
ASD029 1 0
ASD033 1 0
ASD034 1 0
ASD036 1 0
ASD037 1 0
ASD042 1 0
ASD046 1 0
ASD055 1 0
ASD056 1 0
ASD060 1 0
ASD067 1 0
ASD079 1 0
ASD148E 0 1
ASD167C 0 1
ASD168C 0 1
ASD171C 0 1
ASD181C 0 1
ASD184C 0 1
ASD192C 0 1
ASD194C 0 1
ASD200C 0 1
ASD204 0 1
ASD205B 0 1
ASD207B 0 1
ASD300 0 1
ASD301 0 1
ASD302 0 1
ASD303 0 1
ASD304 0 1
ASD305 0 1
ASD306 0 1
ASD307 0 1
ASD308 0 1
ASD4004 0 1
ASD4005 0 1
TD001 1 0
TD004 1 0
TD012 1 0
TD022 1 0
TD029 1 0
TD031 1 0
TD033 1 0
TD035 1 0
TD043 1 0
TD045 1 0
TD051 1 0
TD053 1 0
TD106E 1 0
TD163C 1 0
lop$nSubj vs File:
1
42003 1
42004 1
42007 1
42008 1
42009 1
42010 1
42013 1
42015 1
42016 1
42019 1
42022 1
42023 1
42027 1
42029 1
42030 1
42031 1
42033 1
42035 1
42036 1
42038 1
42039 1
42040 1
42041 1
42042 1
ASD015 1
ASD022 1
ASD029 1
ASD033 1
ASD034 1
ASD036 1
ASD037 1
ASD042 1
ASD046 1
ASD055 1
ASD056 1
ASD060 1
ASD067 1
ASD079 1
ASD148E 1
ASD167C 1
ASD168C 1
ASD171C 1
ASD181C 1
ASD184C 1
ASD192C 1
ASD194C 1
ASD200C 1
ASD204 1
ASD205B 1
ASD207B 1
ASD300 1
ASD301 1
ASD302 1
ASD303 1
ASD304 1
ASD305 1
ASD306 1
ASD307 1
ASD308 1
ASD4004 1
ASD4005 1
TD001 1
TD004 1
TD012 1
TD022 1
TD029 1
TD031 1
TD033 1
TD035 1
TD043 1
TD045 1
TD051 1
TD053 1
TD106E 1
TD163C 1
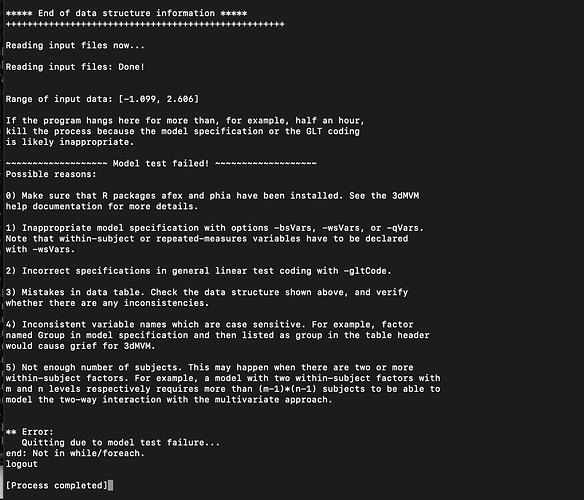
***** End of data structure information *****
++++++++++++++++++++++++++++++++++++++++++++++++++++
Reading input files now...
Reading input files: Done!
Range of input data: [-1.099, 2.606]
If the program hangs here for more than, for example, half an hour,
kill the process because the model specification or the GLT coding
is likely inappropriate.
~~~~~~~~~~~~~~~~~~~ Model test failed! ~~~~~~~~~~~~~~~~~~~
Possible reasons:
0) Make sure that R packages afex and phia have been installed. See the 3dMVM
help documentation for more details.
1) Inappropriate model specification with options -bsVars, -wsVars, or -qVars.
Note that within-subject or repeated-measures variables have to be declared
with -wsVars.
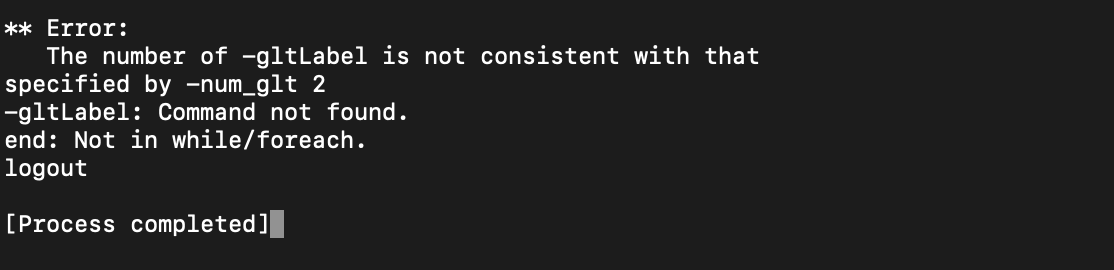
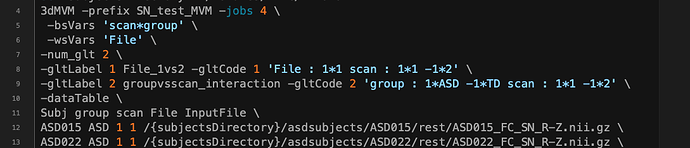
2) Incorrect specifications in general linear test coding with -gltCode.
3) Mistakes in data table. Check the data structure shown above, and verify
whether there are any inconsistencies.
4) Inconsistent variable names which are case sensitive. For example, factor
named Group in model specification and then listed as group in the table header
would cause grief for 3dMVM.
5) Not enough number of subjects. This may happen when there are two or more
within-subject factors. For example, a model with two within-subject factors with
m and n levels respectively requires more than (m-1)*(n-1) subjects to be able to
model the two-way interaction with the multivariate approach.
** Error:
Quitting due to model test failure...
end: Not in while/foreach.
logout
[Process completed]