AFNI version info (afni -ver): 22.3.03
Hi all,
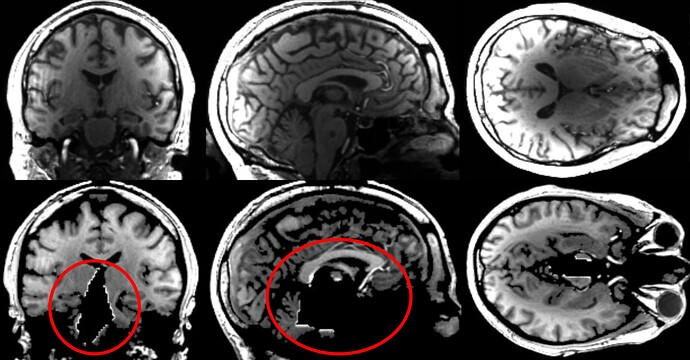
in our study, we collect two T1 images with different inversion times, which I then combine to a computed T1 using the following formula (see 1st row in attached image):
3dcalc -overwrite -a $inv01 -b $inv02 -prefix t1_computed -expr '(a^2)+(b^2)' -float
Afterwards, I subject the T1 to @SSwarper for normalization:
@SSwarper -input t1_computed.nii.gz -base MNI152_2009_template_SSW+tlrc. -subid sswarp_anat2mni -SSopt '-blur_fwhm 2'
However, after 3dunifze (anatU.sswarp_anat2mni.nii), the image looks like in the image bottom row, basically a signal loss in the medial area.
Do you have an idea about what can be done about it?
Thanks,
Philipp
Hi, Philipp-
What if you add the -unifize_off option? That should turn the unifizing off (as advertised!).
Note that there is also an update to @SSwarper, called sswarper2 that has basically the same syntax. You might be interested in trying that out, too. The same -unifize_off option exists.
--pt
Hi Paul,
I did as you recommended and, for testing, just ran the 3dSkullStrip command on the data outside of sswarper (btw sswarper does not exist when i try to call it with @SSwarper2, afni ver: 22.3.03). The result is that now the SkullStrip completely fails and throws an error:
3dSkullStrip -overwrite -input t1_computed.nii.gz -prefix anatSS -debug 1
++ Notice SUMA_BrainWrap_ParseInput (SUMA_3dSkullStrip.c:1244 @21:38:40):
Setting fill_hole to 10
++ Notice 3dSkullStrip (SUMA_3dSkullStrip.c:1600 @21:38:40):
Loading dset, performing Spatial Normalization
*+ WARNING: If you are performing spatial transformations on an oblique dset,
such as t1_computed.nii.gz,
or viewing/combining it with volumes of differing obliquity,
you should consider running:
3dWarp -deoblique
on this and other oblique datasets in the same session.
See 3dWarp -help for details.
++ Oblique dataset: t1_computed.nii.gz is 5.006094 degrees from plumb.
3dSkullStrip:
Original resolution 1.012500, 1.000000, 1.012500
SpatNorm resolution 1.012500, 1.000000, 1.012500
++mri_brainormalize: normalizing
++mri_brainormalize: top clip above slice 153
++mri_brainormalize: Rescaling.
++mri_brainormalize: Eroding, clustering ...
+ Eroded 437033 voxels
+ Restored 410890 eroded voxels
+ Clustering voxels ...
+ Largest cluster has 1660695 voxels
+ Clustering voxels ...
+ Largest cluster has 4533884 voxels
++mri_brainormalize: Clipping extreme values.
+ scaling by fac=0.0271913
3dSkullStrip: -spatnorm: Expecting 6195700 voxels in in_vol dset (167 212 175)
3dSkullStrip: Edge threshold set to 23.545654. (minimum acceptable was 23.545654)
(92.000000 percentile =23.545654, 99.999001 percentile = 235.456543)
3dSkullStrip: Prepping volume...
SUMA_LoadPrepInVol: Volume 1624790.000000, radius 72.929459
3dSkullStrip: Beginning brain extraction...
3dSkullStrip: Have surface, OK for 1st entry.
3dSkullStrip: Stretching to avoid ventricles.
SO->Center is [-0.480866 -4.897505 -4.008005]
SUMA_StretchToFitLeCerveau:
In stage 1
SUMA_StretchToFitLeCerveau:
Stage1: satiated, low MaxExp
SUMA_StretchToFitLeCerveau:
Stage 3, type 1: 0 troubled nodes, going for more...
SUMA_StretchToFitLeCerveau:
In stage 1
SUMA_StretchToFitLeCerveau:
Stage1: satiated, low MaxExp
SUMA_StretchToFitLeCerveau:
Stage 3, type 2: 0 troubled nodes, going for more...
3dSkullStrip: Checking for self intersection...
3dSkullStrip: No intersections found.
3dSkullStrip: Final smoothing of 72
3dSkullStrip: Touchup correction, pass 1 ...
3dSkullStrip: The beauty treatment smoothing.
3dSkullStrip: Locating voxels inside surface ...
Fatal Signal 11 (SIGSEGV) received
SUMA_SurfGridIntersect
SUMA_FindVoxelsInSurface
3dSkullStrip
Bottom of Debug Stack
** AFNI version = AFNI_22.3.03 Compile date = Oct 13 2022
** [[Precompiled binary linux_centos_7_64: Oct 13 2022]]
** Program Death **
Please advise.
Thanks,
Philipp
Hi, Philipp-
Your AFNI version is about 1.5 years old. sswarper2 was added earlier this year, so if you update your version of AFNI, you would have it.
I guess it is possible that for @SSwarper, you might want to add -init_skullstr_off, then. Note that this would be a difference between using sswarper2: you would not add that option there, because this program doesn't try doing and initial skullstrip. It avoids that step.
--pt
Yeah, I was thinking old is gold, but maybe it's time to move on with a new version 
I was actually able to solve the issue. In fact, when I used
3dUnifize -GM t1_computed
the output actually looked good! I have no idea why, but feeding the output to skullstripping also produced a nicely stripped brain!
Running a validation test now within sswarper, which takes more time obviously, but I don't see why it wouldn't work there.
Some things in afni are truly mysterious...
Thanks for your help and until next time!
Philipp