Dear AFNI team,
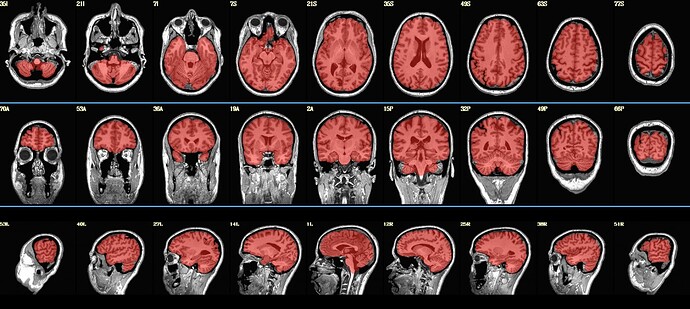
I'm running SSwarper2 on a participant's T1 image (T1.nii.gz) and noticed that the resulting aligned and skull-stripped output appears to exclude most of the cerebellum. I've attached a screenshot showing the issue — the red overlay (presumably the warped template mask) covers the cerebral cortex well, but the cerebellum is largely missing.
I used the default parameters for SSwarper2. The input image looks fine visually and includes the cerebellum before processing.
code:
sswarper2 \
-input T1.nii.gz \
-base MNI152_2009_template_SSW.nii.gz \
-subid D11 \
-odir /to3dfile/SSwarper
Could you advise what might be causing this issue and how I might fix it?
Thank you for your help!
Best,
Zhiqing
Hi, Zhiqing-
Indeed, I see where part (probably not what I'd call "most") of the cerebellum is missing there.
You could try adjusting the final nonlinear cost function, which is "pcl" by default, to "lpa":
sswarper2 \
-input T1.nii.gz \
-base MNI152_2009_template_SSW.nii.gz \
-subid D11 \
-cost_nl_final lpa \
-odir /to3dfile_new/SSwarper
--pt
Thank you so much! I will try it.
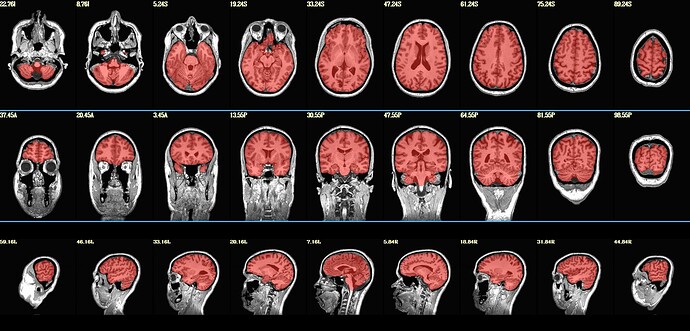
I tried, but still it is not cover all of the cerebellum.
OK, indeed, there is a small amount of cerebellum that is not included in the mask.
Masking in sswarper2 comes from alignment to the template. So, the reason that part of the cerebellum is not included in the mask is that that part is not quite aligned to the template. The amount of warping that can happen is restricted to being diffeomorphic (hence, invertible), and there is also a "stretchiness" factor that is applied within the algorithm so that totally crazy things don't happen. But this is tweakable in 3dQwarp (the workhorse program that sswarper2 wraps around) via a "penalty factor" option called -penfac ...
I have just added control of this option in the last stages of nonlinear alignment within sswarper2, so that you can have access to it if you update your local AFNI. This option is now available within AFNI v25.1.06 and higher. We did a build last night, so you should be able to get it now on any OS.
The option help description here:
-penfac PF :(opt) use the number PF to weight the penalty.
The default value is 1. Larger values of PF mean the
penalty counts more, aiming to reduce grid distortions;
smaller values allow for more flexibility (which can lead
to weird results if too flexible), and the minimum
value is 0. The penalty values specified here are only
applied in the final rounds of 3dQwarp here.
Since the default is 1 and "smaller means stretchier", you could try perhaps adding -penfac 0.5 to your command and see if that helps.
--pt
Thank you for your help and I tried.
It is not good.
Even with -penfac 0.05, it is still not cover all of the cerebellum, but it get better.
This result is what I got with -penfac 0.05: