Hello Masters of AFNI,
I have processing some publicly available rs-fMRI datasets using a pipeline that consists of Freesurfer, SUMA, SSWarper, and afni_proc.py (Example 11). I have run processed a number of datasets using this pipeline but I ran into a fatal error message during the afni_proc.py step with the latest dataset that I have never seen before.
The penultimate error message is:
3dNwarpApply -source copy_af_anat_w_skull+orig -master anat_final.HSC_0020.REST+tlrc -ainterp wsinc5 -nwarp anatQQ.HSC_0020_WARP.nii anatQQ.HSC_0020.aff12.1D -prefix follow_anat_anat_w_skull
++ 3dNwarpApply: AFNI version=AFNI_23.3.13 (Dec 8 2023) [64-bit]
++ Authored by: Zhark the Warped
** ERROR: Can't open -source dataset 'copy_af_anat_w_skull+orig' :-(
** FATAL ERROR: No good source datasets found!
but there are 2 other fatal errors that also occur earlier in the process:
#Script is running (command trimmed):
3dAllineate -nmi -weight ./vr_base_min_outlier_unif_ts_ns_wt+orig -source ./__tt_anatSS.HSC_0020_lr_al_junk+orig -prefix ./anatSS.HSC_0020_flip_al_junk -base ./vr_base_min_outlier_unif_ts_ns+orig -cmass -1Dmatrix_save ./anatSS.HSC_0020_flip__al_junk_mat.aff12.1D -master BASE -mast_dxyz 1.000000 -weight_frac 1.0 -maxrot 6 -maxshf 10 -VERB -warp aff -source_automask+4 -twobest 11 -twopass -VERB -maxrot 45 -maxshf 40 -fineblur 1 -source_automask+2
++ 3dAllineate: AFNI version=AFNI_23.3.13 (Dec 8 2023) [64-bit]
++ Authored by: Zhark the Registrator
** FATAL ERROR: can't open --source dataset './__tt_anatSS.HSC_0020_lr_al_junk+orig' :-(
** Program compile date = Dec 8 2023
#Script is running (command trimmed):
3dAllineate -allcostX1D IDENTITY __tt_lr_flipcosts.1D -weight ./vr_base_min_outlier_unif_ts_ns_wt+orig -source ./anatSS.HSC_0020_flip_al_junk+orig -base ./vr_base_min_outlier_unif_ts_ns+orig -cmass -master BASE -mast_dxyz 1.000000 -weight_frac 1.0 -maxrot 6 -maxshf 10 -VERB -warp aff -source_automask+4 -twobest 11 -twopass -VERB -maxrot 45 -maxshf 40 -fineblur 1 -source_automask+2
++ 3dAllineate: AFNI version=AFNI_23.3.13 (Dec 8 2023) [64-bit]
++ Authored by: Zhark the Registrator
** FATAL ERROR: can't open --source dataset './anatSS.HSC_0020_flip_al_junk+orig' :-(
These errors occur with all of the fMRI volumes included in the dataset but the fMRI volumes themselves seem ok on their surface in that they can be viewed, have the correct number of images, etc.
I have included one example of the afni_proc.py call that I used but the full output log is too large to be attached here. I am not sure if this issue can be identified from what I've provided so please let me know if more information would be helpful. I have tried using both the Haskins atlas and the MNI atlas (for both the SSWarper and afni_proc.py steps) so I don't think that is the issue but that is as far as I was able to get with my troubleshooting.
Thank you for any help you can offer!
Paul
My afni_proc.py script:
afni_proc.py \
-subj_id HSC_0020.REST \
-out_dir /mnt/Illmatic/processed_data/POND32/test/HSC_0020/AFNI \
-blocks despike tshift align tlrc volreg mask scale \
regress \
-radial_correlate_blocks tcat volreg \
-copy_anat /mnt/Illmatic/processed_data/POND32/test/HSC_0020/anatSS.HSC_0020.nii \
-anat_has_skull no \
-anat_follower anat_w_skull anat /mnt/Illmatic/processed_data/POND32/test/HSC_0020/anatU.HSC_0020.nii \
-anat_follower_ROI aaseg anat /mnt/Illmatic/processed_data/POND32/test/HSC_0020/SUMA/aparc.a2009s+aseg_REN_all.nii \
-anat_follower_ROI aeseg epi /mnt/Illmatic/processed_data/POND32/test/HSC_0020/SUMA/aparc.a2009s+aseg_REN_all.nii \
-anat_follower_ROI FSvent epi /mnt/Illmatic/processed_data/POND32/test/HSC_0020/SUMA/fs_ap_latvent.nii.gz \
-anat_follower_ROI FSWe epi /mnt/Illmatic/processed_data/POND32/test/HSC_0020/SUMA/fs_ap_wm.nii.gz \
-anat_follower_erode FSvent FSWe \
-dsets /mnt/Illmatic/processed_data/POND32/test/HSC_0020/raw_data/PND03_HSC_0020_01_SE01_MR_12-fMRI_RSN.nii \
-tcat_remove_first_trs 2 \
-align_unifize_epi local \
-align_opts_aea -cost nmi -giant_move -check_flip \
-tlrc_base HaskinsPeds_NL_template1.0_SSW.nii.gz \
-tlrc_NL_warp \
-tlrc_NL_warped_dsets /mnt/Illmatic/processed_data/POND32/test/HSC_0020/anatQQ.HSC_0020.nii /mnt/Illmatic/processed_data/POND32/test/HSC_0020/anatQQ.HSC_0020.aff12.1D \
/mnt/Illmatic/processed_data/POND32/test/HSC_0020/anatQQ.HSC_0020_WARP.nii \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-mask_apply anat \
-regress_motion_per_run \
-regress_ROI_PC FSvent 3 \
-regress_ROI_PC_per_run FSvent \
-regress_make_corr_vols aeseg FSvent \
-regress_anaticor_fast \
-regress_anaticor_label FSWe \
-regress_censor_motion 0.4 \
-regress_censor_outliers 0.05 \
-regress_apply_mot_types demean deriv \
-regress_est_blur_epits \
-regress_est_blur_errts \
-html_review_style pythonic
Hi, Paul-
I wonder if this new dataset has the coordinates of the anatomical being very far away from the template dataset, and therefore an intermediate warp dataset is blowing up in size and not able to be created.
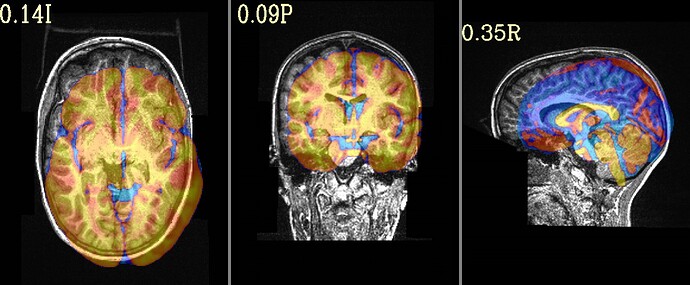
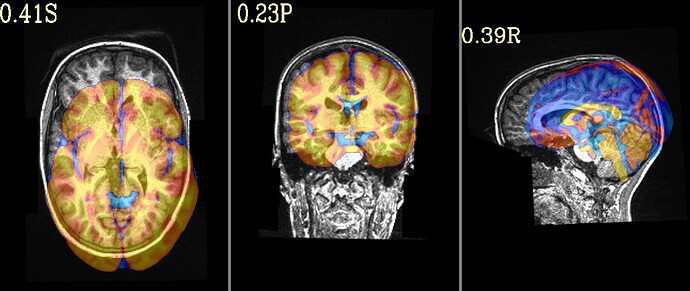
To test that, could you post what the image (or images) created by this command are:
@djunct_overlap_check \
-ulay DSET_ANAT \
-olay HaskinsPeds_NL_template1.0_SSW.nii.gz'[0]' \
-prefix img_olap
... where DSET_ANAT is the dataset you put into @SSwarper?
thanks,
pt
ps: here is your AP command vertically spaced out, with a variable to shorten the path, in case that is helpful for reading/working with, either for this MB thread or for your general workflow:
set dir_data = /mnt/Illmatic/processed_data/POND32/test/HSC_0020
afni_proc.py \
-subj_id HSC_0020.REST \
-out_dir ${dir_data}/AFNI \
-blocks despike tshift align tlrc volreg mask scale \
regress \
-radial_correlate_blocks tcat volreg \
-copy_anat ${dir_data}/anatSS.HSC_0020.nii \
-anat_has_skull no \
-anat_follower anat_w_skull anat \
${dir_data}/anatU.HSC_0020.nii \
-anat_follower_ROI aaseg anat \
${dir_data}/SUMA/aparc.a2009s+aseg_REN_all.nii \
-anat_follower_ROI aeseg epi \
${dir_data}/SUMA/aparc.a2009s+aseg_REN_all.nii \
-anat_follower_ROI FSvent epi \
${dir_data}/SUMA/fs_ap_latvent.nii.gz \
-anat_follower_ROI FSWe epi ${dir_data}/SUMA/fs_ap_wm.nii.gz \
-anat_follower_erode FSvent FSWe \
-dsets ${dir_data}/raw_data/PND03_HSC_0020_01_SE01_MR_12-fMRI_RSN.nii \
-tcat_remove_first_trs 2 \
-align_unifize_epi local \
-align_opts_aea -cost nmi \
-giant_move \
-check_flip \
-tlrc_base HaskinsPeds_NL_template1.0_SSW.nii.gz \
-tlrc_NL_warp \
-tlrc_NL_warped_dsets ${dir_data}/anatQQ.HSC_0020.nii \
${dir_data}/anatQQ.HSC_0020.aff12.1D \
${dir_data}/anatQQ.HSC_0020_WARP.nii \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-mask_apply anat \
-regress_motion_per_run \
-regress_ROI_PC FSvent 3 \
-regress_ROI_PC_per_run FSvent \
-regress_make_corr_vols aeseg FSvent \
-regress_anaticor_fast \
-regress_anaticor_label FSWe \
-regress_censor_motion 0.4 \
-regress_censor_outliers 0.05 \
-regress_apply_mot_types demean deriv \
-regress_est_blur_epits \
-regress_est_blur_errts \
-html_review_style pythonic
Hi Paul,
Thanks for your suggestion. I have modified the code you included and have attached the images you requested. The overlap doesn't look too bad to me but I defer to your expertise. What would you suggest as the next steps?
Thank you for the formatting lesson! That looks much cleaner than my version :)
Paul
Hi, Paul-
Thanks for posting that. Indeed, the overlap does look good, so that is nice. So, if that is not the issue, I wonder if the sform_code and qform_code values of the anatomical are annoying ambiguous. Presuming the dset you put into @SSwarper is a NIFTI dset, what is the output of:
nifti_tool \
-disp_hdr -field qform_code -field sform_code \
-infiles DSET_ANAT
? Or, if it is a BRIK/HEAD file, what is its name?
One other thing I notice from this output is that your anatomical dataset has obliquity in it. The way I can tell is that 2 images are output, and one has "DEOB" in the name---that is what the overlap snapshot program will do if either the overlay or underlay dataset has obliquity (and in this case we know that the Haskins reference template does not have any).
Obliquity is usually something we try to remove from the anatomical before processing (even before @SSwarper, and also before FreeSurfer's recon-all, if you are using that). Even though there isn't a large amount, it is something that different software deal with differently (so, FreeSurfer will apply it while most AFNI alignment programs tend to ignore it), and we want to avoid that mismatch. This post describes 2 ways to remove the obliquity from the anatomical while preserving the coordinate origin (x,y,z)=(0,0,0) and also not smoothing/regridding the data at all.
--pt
Hi Paul,
Here are the sform/qform results.
name offset nvals values
qform_code 252 1 0
sform_code 254 1 2
From my brief reading (and limited understanding), it appears as though the 2 in the sform_code could be the problem.
I will deoblique my structural images in the meantime.
Thank you once again for your help in sorting this out.
Paul
Ahhhhh, the dreaded 2 code! Yes, it is.
What is therefore likely happening is that when that dataset is copied to BRIK/HEAD format on input, it gets a +tlrc view-space extension, rather than +orig. Hence, the issue with the proc script looking for 'copy_af_anat_w_skull+orig', but through the sform_code trickery the actual file is called 'copy_af_anat_w_skull+tlrc'. The resulting mismatch is causing the error message woe.
So, I would copy the initial (unprocessed here) anatomical and fix the qform_code and sform_code in it to be something appropriate for original/native space data. At the same time, I would also deoblique the anatomical. In total, that could mean running (now updated to add in a correction mentioned by @pmetzak):
# purge any extensions (NB: "ALL" is a keyword to leave unchanged here)
nifti_tool \
-rm_ext ALL \
-prefix __tmp1.nii \
-infiles DSET_ANAT
# reset [qs]form_code values
nifti_tool \
-mod_hdr \
-mod_field qform_code 1 \
-mod_field sform_code 1 \
-prefix __tmp2.nii \
-infiles __tmp1.nii
# copy to BRIK/HEAD format temporarily and deoblique around origin (w/o resampling)
3dcopy __tmp2.nii __tmp3
3drefit -oblique_recenter __tmp3+orig
3drefit -deoblique __tmp3+orig
3dcopy __tmp3+orig DSET_ANAT_NEW.nii.gz
... and check what you have by running:
3dinfo -obliquity -space DSET_ANAT_NEW.nii.gz
... and the output should look like 0.000 ORIG.
Then, you should be all good to put DSET_ANAT_NEW.nii.gz into @SSwarper (and FreeSurfer, if so processing), and then carry on into afni_proc.py.
--pt
Hi Paul,
Thanks for the providing the commands to correct the qform/sform value issue. However, I did run into an error with the first nifti_tool command when I tried correcting the unprocessed anatomical files. I noticed that someone else ran into a similar error but their issue was with the qform/sform parameters in their functional data so I'm not quite sure how to proceed. The error message can be found below.
Thanks again for your assistance!
Paul
** cannot remove 1 exts from image 'PND03_HBK_0517_01_SE01_MR_T1_deID.nii.gz' with only 0
** ERROR (nifti_read_n1_hdr): failed to find header file for '__tmp1.nii'
Hi, Paul-
I guess that means that there are no extensions in that file to remove. Rather than just copying the file and mentioning that there are none, it is making an error output.
So, you can do the following---but note that another quirk of using nifti_tool to modify header information is that the DSET_ANAT cannot be gzipped for the first step. So, of DSET_ANAT is *.nii.gz, you will first have to run gunzip DSET_ANAT on it, then continue processing as described here (now updated to add in a correction mentioned by @pmetzak):
# reset [qs]form_code values
nifti_tool \
-mod_hdr \
-mod_field qform_code 1 \
-mod_field sform_code 1 \
-prefix __tmp2.nii \
-infiles DSET_ANAT
# copy to BRIK/HEAD format temporarily and deoblique around origin (w/o resampling)
3dcopy __tmp2.nii __tmp3
3drefit -oblique_recenter __tmp3+orig
3drefit -deoblique __tmp3+orig
3dcopy __tmp3+orig DSET_ANAT_NEW.nii.gz
--pt
Thanks again Paul, I was able to use this code to fix the qform/sform problem. If anyone else runs into a similar issue, I'll just note that I needed to make 2 minor corrections to get this to work.
-
On line 6, I removed the .gz from the prefix filename as nifti_tool was unable to reopen the compressed __tmp2 file after it was created and therefore could not modify the qform/sform values.
-
On line 11, I corrected a minor typo and added a dash before oblique_recenter.
Thank you once again for all your help, it would have been very difficult for me to diagnose/correct this issue without your assistance!
Hi, Paul-
Thanks for mentioning those points. I've now edited the commands above fixing those items, so future generations who excitedly grab the code snippets to run them without reading all the down the thread will not suffer so greatly.
Happy processing from here!
--pt