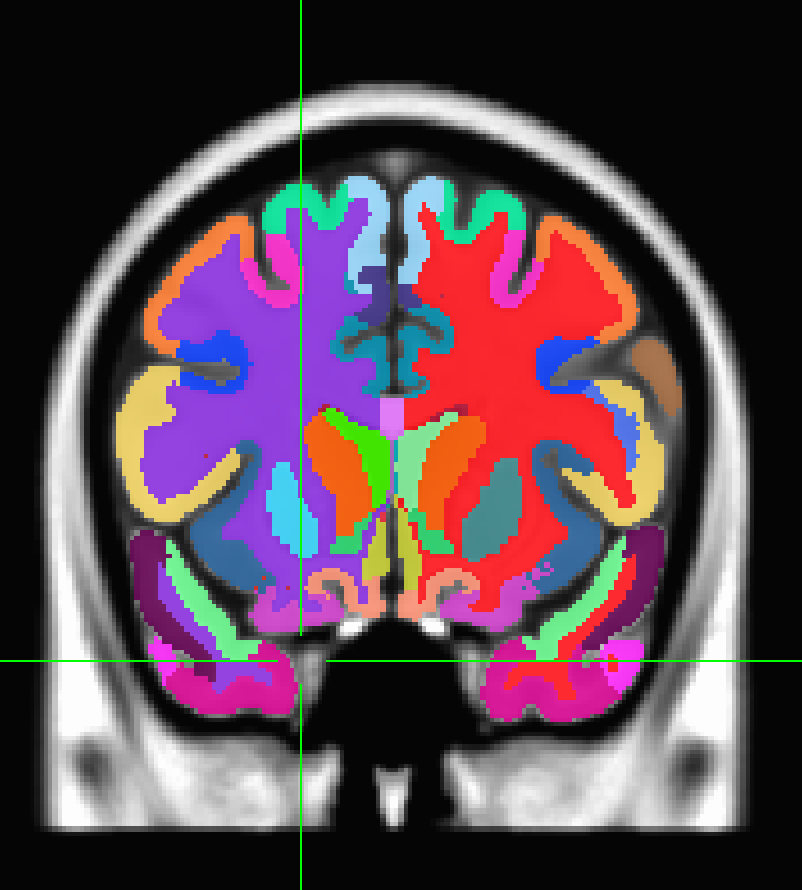
Hello,
I have a segmentation NIfTI file (aparc+aseg+vep.nii) and a LUT file (VEP_atlas_AFNI.lt) that mimics the AFNI .lt format as follows,
<VALUE_LABEL_DTABLE
ni_type="2*String"
ni_dimen="200"
pbar_name="VEP_atlas">
"0" "Unknown"
"1" "Left-Cerebral-Exterior"
"2" "Left-Cerebral-White-Matter"
"3" "Left-Cerebral-Cortex"
"4" "Left-Lateral-Ventricle"
"5" "Left-Inf-Lat-Vent"
"6" "Left-Cerebellum-Exterior"
"7" "Left-Cerebellum-White-Matter"
"8" "Left-Cerebellum-Cortex"
"9" "Left-Thalamus"
"10" "Left-Thalamus-Proper"
"11" "Left-Caudate"
"12" "Left-Putamen"
"13" "Left-Pallidum"
"14" "3rd-Ventricle"
"15" "4th-Ventricle"
"16" "Brain-Stem"
"17" "Left-Hippocampus"
"18" "Left-Amygdala"
"19" "Left-Insula"
"20" "Left-Operculum"
"21" "Line-1"
"22" "Line-2"
"23" "Line-3"
"24" "CSF"
"25" "Left-Lesion"
"26" "Left-Accumbens-area"
"27" "Left-Substancia-Nigra"
"28" "Left-VentralDC"
"29" "Left-undetermined"
"30" "Left-vessel"
"31" "Left-choroid-plexus"
"32" "Left-F3orb"
"33" "Left-lOg"
"34" "Left-aOg"
"35" "Left-mOg"
"36" "Left-pOg"
"37" "Left-Stellate"
"38" "Left-Porg"
"39" "Left-Aorg"
"40" "Right-Cerebral-Exterior"
"41" "Right-Cerebral-White-Matter"
"42" "Right-Cerebral-Cortex"
"43" "Right-Lateral-Ventricle"
"44" "Right-Inf-Lat-Vent"
"45" "Right-Cerebellum-Exterior"
"46" "Right-Cerebellum-White-Matter"
"47" "Right-Cerebellum-Cortex"
"48" "Right-Thalamus"
"49" "Right-Thalamus-Proper"
"50" "Right-Caudate"
"51" "Right-Putamen"
"52" "Right-Pallidum"
"53" "Right-Hippocampus"
"54" "Right-Amygdala"
"55" "Right-Insula"
"56" "Right-Operculum"
"57" "Right-Lesion"
"58" "Right-Accumbens-area"
"59" "Right-Substancia-Nigra"
"60" "Right-VentralDC"
"61" "Right-undetermined"
"62" "Right-vessel"
"63" "Right-choroid-plexus"
"64" "Right-F3orb"
"65" "Right-lOg"
"66" "Right-aOg"
"67" "Right-mOg"
"68" "Right-pOg"
"69" "Right-Stellate"
"70" "Right-Porg"
"71" "Right-Aorg"
"72" "5th-Ventricle"
"73" "Left-Interior"
"74" "Right-Interior"
"77" "WM-hypointensities"
"78" "Left-WM-hypointensities"
"79" "Right-WM-hypointensities"
"80" "non-WM-hypointensities"
"81" "Left-non-WM-hypointensities"
"82" "Right-non-WM-hypointensities"
"83" "Left-F1"
"84" "Right-F1"
"85" "Optic-Chiasm"
"192" "Corpus_Callosum"
"251" "CC_Posterior"
"252" "CC_Mid_Posterior"
"253" "CC_Central"
"254" "CC_Mid_Anterior"
"255" "CC_Anterior"
"1000" "ctx-lh-unknown"
"2000" "ctx-rh-unknown"
"71001" "Left-Frontal-pole"
"71002" "Left-Orbito-frontal-cortex"
"71003" "Left-Gyrus-rectus"
"71004" "Left-F3-Pars-Orbitalis"
"71005" "Left-F3-Pars-triangularis"
"71006" "Left-F3-pars-opercularis"
"71007" "Left-Inferior-frontal-sulcus"
"71008" "Left-F2-rostral"
"71009" "Left-F2-caudal"
"71010" "Left-Middle-frontal-sulcus"
"71011" "Left-SFS-rostral"
"71012" "Left-SFS-caudal"
"71013" "Left-F1-mesial-prefrontal"
"71014" "Left-PreSMA"
"71015" "Left-SMA"
"71016" "Left-F1-lateral-prefrontal"
"71017" "Left-F1-lateral-premotor"
"71018" "Left-Subcallosal-area"
"71019" "Left-Precentral-sulcus-inferior-part"
"71020" "Left-Precentral-sulcus-superior-part"
"71021" "Left-Precentral-gyrus-head-face"
"71022" "Left-Precentral-gyrus-upper-limb"
"71023" "Left-Central-sulcus-head-face"
"71024" "Left-Central-sulcus-upper-limb"
"71025" "Left-Paracentral-lobule"
"71026" "Left-Central-operculum"
"71027" "Left-Parietal-operculum"
"71028" "Left-Anterior-cingulate-cortex"
"71029" "Left-Middle-cingulate-cortex-anterior-part"
"71030" "Left-Middle-cingulate-cortex-posterior-part"
"71031" "Left-Posterior-cingulate-cortex-dorsal"
"71032" "Left-Posterior-cingulate-cortex-retrosplenial-gyrus"
"71033" "Left-Insula-gyri-brevi"
"71034" "Left-Insula-gyri-longi"
"71035" "Left-Temporal-pole"
"71036" "Left-T1-planum-polare"
"71037" "Left-Gyrus-of-Heschl"
"71038" "Left-T1-planum-temporale"
"71039" "Left-T1-lateral-anterior"
"71040" "Left-T1-lateral-posterior"
"71041" "Left-STS-anterior"
"71042" "Left-STS-posterior"
"71043" "Left-ITS-anterior"
"71044" "Left-ITS-posterior"
"71045" "Left-T2-anterior"
"71046" "Left-T2-posterior"
"71047" "Left-T3-anterior"
"71048" "Left-T3-posterior"
"71049" "Left-Fusiform-gyrus"
"71050" "Left-Occipito-temporal-sulcus"
"71051" "Left-Collateral-sulcus"
"71052" "Left-Lingual-sulcus"
"71053" "Left-Parahippocampal-cortex"
"71054" "Left-Rhinal-cortex"
"71055" "Left-Postcentral-gyrus"
"71056" "Left-Postcentral-sulcus"
"71057" "Left-Superior-parietal-lobule-P1"
"71058" "Left-Supramarginal-anterior"
"71059" "Left-Supramarginal-posterior"
"71060" "Left-Angular-gyrus"
"71061" "Left-Intraparietal-sulcus"
"71062" "Left-Precuneus"
"71063" "Left-Marginal-branch-of-the-cingulate-sulcus"
"71064" "Left-Parieto-occipital-sulcus"
"71065" "Left-Anterior-occipital-sulcus-and-preoccipital-notch"
"71066" "Left-O3"
"71067" "Left-O2"
"71068" "Left-O1"
"71069" "Left-Occipital-pole"
"71070" "Left-Lingual-gyrus"
"71071" "Left-Calcarine-sulcus"
"71072" "Left-Cuneus"
"71073" "Left-Hippocampus-anterior"
"71074" "Left-Hippocampus-posterior"
"71075" "Left-Caudate-nucleus"
"71076" "Left-Nucleus-accumbens"
"71077" "Left-Cerebellar-cortex"
"72001" "Right-Frontal-pole"
"72002" "Right-Orbito-frontal-cortex"
"72003" "Right-Gyrus-rectus"
"72004" "Right-F3-Pars-Orbitalis"
"72005" "Right-F3-Pars-triangularis"
"72006" "Right-F3-pars-opercularis"
"72007" "Right-Inferior-frontal-sulcus"
"72008" "Right-F2-rostral"
"72009" "Right-F2-caudal"
"72010" "Right-Middle-frontal-sulcus"
"72011" "Right-SFS-rostral"
"72012" "Right-SFS-caudal"
"72013" "Right-F1-mesial-prefrontal"
"72014" "Right-PreSMA"
"72015" "Right-SMA"
"72016" "Right-F1-lateral-prefrontal"
"72017" "Right-F1-lateral-premotor"
"72018" "Right-Subcallosal-area"
"72019" "Right-Precentral-sulcus-inferior-part"
"72020" "Right-Precentral-sulcus-superior-part"
"72021" "Right-Precentral-gyrus-head-face"
"72022" "Right-Precentral-gyrus-upper-limb"
"72023" "Right-Central-sulcus-head-face"
"72024" "Right-Central-sulcus-upper-limb"
"72025" "Right-Paracentral-lobule"
"72026" "Right-Central-operculum"
"72027" "Right-Parietal-operculum"
"72028" "Right-Anterior-cingulate-cortex"
"72029" "Right-Middle-cingulate-cortex-anterior-part"
"72030" "Right-Middle-cingulate-cortex-posterior-part"
"72031" "Right-Posterior-cingulate-cortex-dorsal"
"72032" "Right-Posterior-cingulate-cortex-retrosplenial-gyrus"
"72033" "Right-Insula-gyri-brevi"
"72034" "Right-Insula-gyri-longi"
"72035" "Right-Temporal-pole"
"72036" "Right-T1-planum-polare"
"72037" "Right-Gyrus-of-Heschl"
"72038" "Right-T1-planum-temporale"
"72039" "Right-T1-lateral-anterior"
"72040" "Right-T1-lateral-posterior"
"72041" "Right-STS-anterior"
"72042" "Right-STS-posterior"
"72043" "Right-ITS-anterior"
"72044" "Right-ITS-posterior"
"72045" "Right-T2-anterior"
"72046" "Right-T2-posterior"
"72047" "Right-T3-anterior"
"72048" "Right-T3-posterior"
"72049" "Right-Fusiform-gyrus"
"72050" "Right-Occipito-temporal-sulcus"
"72051" "Right-Collateral-sulcus"
"72052" "Right-Lingual-sulcus"
"72053" "Right-Parahippocampal-cortex"
"72054" "Right-Rhinal-cortex"
"72055" "Right-Postcentral-gyrus"
"72056" "Right-Postcentral-sulcus"
"72057" "Right-Superior-parietal-lobule-P1"
"72058" "Right-Supramarginal-anterior"
"72059" "Right-Supramarginal-posterior"
"72060" "Right-Angular-gyrus"
"72061" "Right-Intraparietal-sulcus"
"72062" "Right-Precuneus"
"72063" "Right-Marginal-branch-of-the-cingulate-sulcus"
"72064" "Right-Parieto-occipital-sulcus"
"72065" "Right-Anterior-occipital-sulcus-and-preoccipital-notch"
"72066" "Right-O3"
"72067" "Right-O2"
"72068" "Right-O1"
"72069" "Right-Occipital-pole"
"72070" "Right-Lingual-gyrus"
"72071" "Right-Calcarine-sulcus"
"72072" "Right-Cuneus"
"72073" "Right-Hippocampus-anterior"
"72074" "Right-Hippocampus-posterior"
"72075" "Right-Caudate-nucleus"
"72076" "Right-Nucleus-accumbens"
"72077" "Right-Cerebellar-cortex"
</VALUE_LABEL_DTABLE>
I added it to the NIFTI header using
3drefit -labeltable VEP_atlas_AFNI.lt aparc+aseg+vep.nii
the parcel names are displayed correctly, but the there are only two colors shown, one for grey and the other for white matter.
I tried MakeColorMap -in VepFreeSurferColorLutClean.txt -prefix aparc+aseg+vep with VepFreeSurferColorLutClean.txt looking like
#No. Label Name: R G B A
0 Unknown 0 0 0 0
1 Left-Cerebral-Exterior 70 130 180 0
2 Left-Cerebral-White-Matter 245 245 245 0
3 Left-Cerebral-Cortex 205 62 78 0
4 Left-Lateral-Ventricle 120 18 134 0
5 Left-Inf-Lat-Vent 196 58 250 0
6 Left-Cerebellum-Exterior 0 148 0 0
7 Left-Cerebellum-White-Matter 220 248 164 0
8 Left-Cerebellum-Cortex 230 148 34 0
9 Left-Thalamus 0 118 14 0
10 Left-Thalamus-Proper 0 118 14 0
11 Left-Caudate 122 186 220 0
12 Left-Putamen 236 13 176 0
13 Left-Pallidum 12 48 255 0
14 3rd-Ventricle 204 182 142 0
15 4th-Ventricle 42 204 164 0
16 Brain-Stem 119 159 176 0
17 Left-Hippocampus 220 216 20 0
...
I placed the segmentation file and the corresponding nimal.cmap file (with the same name) in the same folder, but the colors in AFNI did not update. Is there a way to make the AFNI GUI automatically display the LUT colors?
Thanks