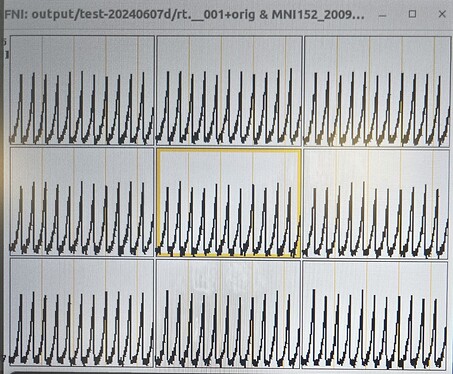
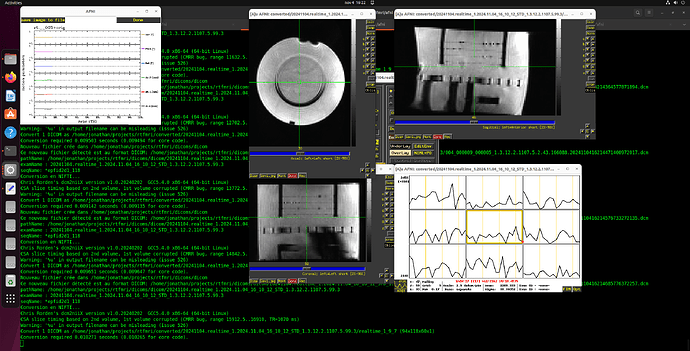
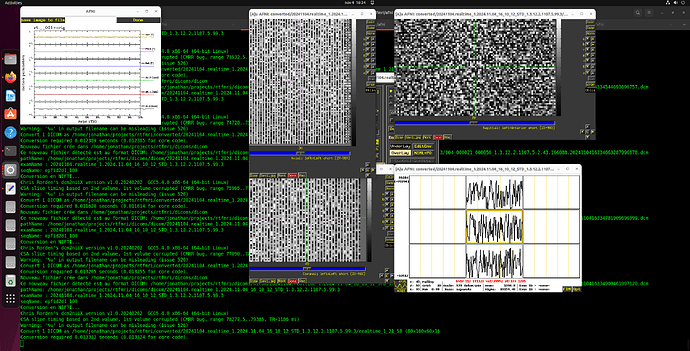
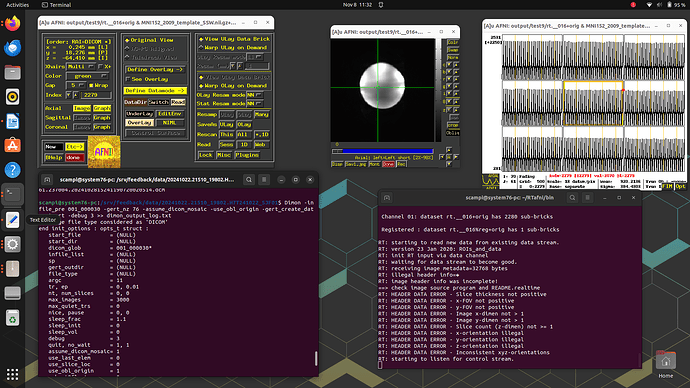
I did more troubleshooting -- I think the issue is that each volume is being interpreted in 2D rather than 3D. You can see this in the output data below and on the coronal and saggittal slices of the output image (which are a single band of 1 voxel depth.
I'm assuming this is a DICOM header issue, but I am not aware of any way to change it-- FYI I double checked and we are currently using XA60 on the scanner. I am not sure why specifying the -gert_nz is not fixing this issue by clarifying that there are 76 slices. However, I have tried every option in the Dimon documentation and am not seeing any change in behavior. (Changing the "Update" option in the RT plugin did fix the lag issue.)
Is there any way to tell Dimon the "illegal header info" manually? (i.e., specifying the x-dimen and y-dimen and fov variables?)
Below I've included the output from running it the Dimon command you mentioned, with the debug turned up to 3. I have also taken a screenshot of the output in AFNI itself so you can have a better sense of the issue (you'll notice there are no Saggital/Coronal images available since there is no data in that dimension).
Thank you for your continued help or any other insight you might have
Output on afni -rt terminal
RT: waiting for data stream to become good.
RT: receiving image metadata=32768 bytes
RT: illegal header info=�
RT: image header info was incomplete!
==> check image source program and README.realtime
RT: HEADER DATA ERROR - Slice thickness not positive
RT: HEADER DATA ERROR - x-FOV not positive
RT: HEADER DATA ERROR - y-FOV not positive
RT: HEADER DATA ERROR - Image x-dimen not > 1
RT: HEADER DATA ERROR - Image y-dimen not > 1
RT: HEADER DATA ERROR - Slice count (z-dimen) not >= 1
RT: HEADER DATA ERROR - x-orientation illegal
RT: HEADER DATA ERROR - y-orientation illegal
RT: HEADER DATA ERROR - z-orientation illegal
RT: HEADER DATA ERROR - Inconsistent xyz-orientations
RT: starting to listen for control stream.
Dimon terminal
Dimon -infile_pre 001_000030 -gert_nz 76 -assume_dicom_mosaic -use_obl_origin -gert_create_dataset -rt -debug 3
-- image file type considered as 'DICOM'
end init_options : opts_t struct :
start_file = (NULL)
start_dir = (NULL)
dicom_glob = 001_000030*
infile_list = (NULL)
sp = (NULL)
gert_outdir = (NULL)
file_type = (NULL)
argc = 11
tr, ep = 0, 0.01
nt, num_slices = 0, 0
max_images = 3000
max_quiet_trs = 0
nice, pause = 0, 0
sleep_frac = 1.1
sleep_init = 0
sleep_vol = 0
debug = 3
quit, no_wait = 1, 1
assume_dicom_mosaic= 1
use_last_elem = 0
use_slice_loc = 0
use_obl_origin = 1
ushort2float = 0
show_sorted_list = 0
gert_reco = 1
gert_filename = (NULL)
gert_prefix = (NULL)
chan_digits = 3
chan_prefix = (NULL)
gert_nz = 76
gert_format = 0
gert_exec = 1
gert_quiterr = 0
dicom_org = 0
sort_num_suff = 0
sort_acq_time = 0
order_as_zt = 0
read_all = 1
rev_org_dir = 0
rev_sort_dir = 0
save_errors = 0
flist_file = (NULL)
flist_details = (NULL)
sort_method = (NULL)
(rt, swap, rev_bo) = (1, 0, 0)
num_chan = 0
host = (NULL)
drive_list(u,a,p) = 0, 0, (nil)
wait_list (u,a,p) = 0, 0, (nil)
rt_list (u,a,p) = 0, 0, (nil)
end init_options : param_t struct :
ftype = 4
glob_dir = 001_000030*
fnames_done = 0
nfim = 0
nfalloc = 0
fim_update = 0
fim_skip = 0
fim_start = 0
max2read = -1
Dimon version 4.34 (September 5, 2024) running, use to quit...
ART: starting I/O to afni
ART: Opening control channel tcp:localhost:7961 to AFNI.
ART: Entering AFNI_WAIT_CONTROL_MODE.
ART: Control channel connected to AFNI. Entering AFNI_OPEN_DATA_MODE.
ART: Sending control information to AFNI:
tcp:localhost:7960
ART: Opening data channel tcp:localhost:7960 to AFNI.
Can't connect? tcp_connect[connect]: Connection refused
ART: Entering AFNI_CATCHUP_MODE.
ART: Entering AFNI_CONTINUE_MODE.
ART: comm link to afni established at
-- scanning for first volume
-- sorted fnames: from 0 to 30
-- appended 30 fnames to fim_o
-- not a Siemens Mosaic
-- used process_siemens_mosaic...
++ Data detected to be oblique
-- is_oblique = 1, im_is_volume = 0
** CID: have non-mosaic 001_000030_000001_1.3.12.2.1107.5.2.61.237004.2024102815241190720020514.dcm with 76 images
-- not a Siemens Mosaic
-- used process_siemens_mosaic...
*+ WARNING: Bad DICOM header - assuming oblique scaling direction!
post mri_read_dicom_get_obliquity -- oblique info = 2
1.5914 0.1650 -0.0184 -111.7550
-0.1660 1.5856 -0.1353 -79.3235
0.0042 0.1365 1.5942 -64.4097
0.0000 0.0000 0.0000 1.0000
** CID: have non-mosaic 001_000030_000002_1.3.12.2.1107.5.2.61.237004.2024102815241266975020561.dcm with 76 images
no more warnings will be printed...
-- RNI: read 30 new images (nerrs = 0)
-- fim_o: sorting image list via method UNSPECIFIED
(30 images from 0 to 29)
-- read_image_files: 30 images, start = 0, proc = 30, tot = 30
-- VS state 0, bound 30, prev -1, first 0, pf -1
+d found single slice volume
-- data is oblique
-- volume search returns 1
-- first volume found (1 slices)
+d first volume : vol_t struct at :
nim = 1
fs_1 = 0
first_file = 001_000030_000001_1.3.12.2.1107.5.2.61.237004.2024102815241190720020514.dcm
last_file = 001_000030_000001_1.3.12.2.1107.5.2.61.237004.2024102815241190720020514.dcm
(z_first, z_last) = (-64.4097, -64.4097)
z_delta, image_dz = (1, 1.6)
oblique = 1
(seq_num, run) = (-1, 30)
+d first volume : ge_header_info at :
good = 1
(nx,ny) = (130,130)
uv17 = 30
index = 1
im_index = -1
atime = 0.000000
slice_loc = 0.000000
(dx,dy,dz) = (1.6,1.6,1.6)
zoff = -64.4097
(tr,te) = (1,0)
orients = RLAPIS
+d first volume : ge_extras :
bpp = 2
cflag = 0
hdroff = -1
skip = -1
swap = 0
kk = 0
ge_echo_time = -1
ge_echo_num = 1
ge_me_index = -1
ge_nim_acq = -1
sop_iuid_maj = 237004
sop_iuid_min = -1
xorg = -111.755
yorg = -79.3235
(xyz0,xyz1,xyz2) = (0,0,0)
(xyz3,xyz4,xyz5) = (0,0,0)
(xyz6,xyz7,xyz8) = (0,0,0)
+d first volume : mosaic_info :
im_is_volume = 0
nslices = 0
mos_nx, ny = 0, 0
completing orients from 'RLAPIS' to 'RLAPIS'
-- system order is LSB_FIRST, image order is LSB_FIRST
ART: dataset control info (718 bytes) to afni:
ACQUISITION_TYPE 3D+timing
ZORDER seq
TR 1.000000
XYFOV 208.000000 208.000000 1.600000
XYMATRIX 130 130 1
DATUM short
XYZAXES R-L A-P I-S
XYZFIRST 111.754997R 79.323502A 64.409698I
BYTEORDER LSB_FIRST
OBLIQUE_XFORM 1.591362 0.165013 -0.018357 -111.754997 -0.165976 1.585603 -0.135331 -79.323502 0.004235 0.136505 1.594161 -64.409698 0.000000 0.000000 0.000000 1.000000
DRIVE_AFNI OPEN_WINDOW axialimage
DRIVE_AFNI OPEN_WINDOW axialgraph
NOTE created remotely via real-time afni
starting with file : '001_000030_000001_1.3.12.2.1107.5.2.61.237004.2024102815241190720020514.dcm'
creation command : Dimon -infile_pre 001_000030 -gert_nz 76 -assume_dicom_mosaic -use_obl_origin -gert_create_dataset -rt -debug 3
ART: control info sent OK, rv = 719
ART: sent images from volume (30:1) to host localhost
-d first vol - new params : param_t struct :
ftype = 4
glob_dir = 001_000030*
fnames_done = 0
nfim = 30
nfalloc = 50
fim_update = 1
fim_skip = 0
fim_start = 1
max2read = -1
-d ftype: DICOM (4)
-- first vol ART_comm struct :
(state, mode) = (3, 5)
(use_tcp, swap) = (1, 0)
byte_order = 1
is_oblique = 1
zorder = (NULL)
host = localhost
ioc_name = tcp:localhost:7960
oblique_xform:
1.5914 0.1650 -0.0184 -111.7550
-0.1660 1.5856 -0.1353 -79.3235
0.0042 0.1365 1.5942 -64.4097
0.0000 0.0000 0.0000 1.0000
-d computed nap_time is 1100 ms (TR = 1.00)
-- scanning for additional volumes...
-- run 30: 1 ++ nap time = 1100, tr_naps = 1 (max quiet time 2.200s)
-- svs: init alloc - vol 1, run 30, file 001_000030_000001_1.3.12.2.1107.5.2.61.237004.2024102815241190720020514.dcm
-- svs: realloc (50 entries) - vol 1, run 30, file 001_000030_000001_1.3.12.2.1107.5.2.61.237004.2024102815241190720020514.dcm
-- svs: run 30, seq_num 1
++ volume match: have volume from index 1 to 1
-- vol match returns 1
2
-- svs: run 30, seq_num 2
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:2) to host localhost
++ volume match: have volume from index 2 to 2
-- vol match returns 1
3
-- svs: run 30, seq_num 3
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:3) to host localhost
++ volume match: have volume from index 3 to 3
-- vol match returns 1
4
-- svs: run 30, seq_num 4
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:4) to host localhost
++ volume match: have volume from index 4 to 4
-- vol match returns 1
5
-- svs: run 30, seq_num 5
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:5) to host localhost
++ volume match: have volume from index 5 to 5
-- vol match returns 1
6
-- svs: run 30, seq_num 6
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:6) to host localhost
++ volume match: have volume from index 6 to 6
-- vol match returns 1
7
-- svs: run 30, seq_num 7
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:7) to host localhost
++ volume match: have volume from index 7 to 7
-- vol match returns 1
8
-- svs: run 30, seq_num 8
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:8) to host localhost
++ volume match: have volume from index 8 to 8
-- vol match returns 1
9
-- svs: run 30, seq_num 9
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:9) to host localhost
++ volume match: have volume from index 9 to 9
-- vol match returns 1
10
-- svs: run 30, seq_num 10
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:10) to host localhost
++ volume match: have volume from index 10 to 10
-- vol match returns 1
11
-- svs: run 30, seq_num 11
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:11) to host localhost
++ volume match: have volume from index 11 to 11
-- vol match returns 1
12
-- svs: run 30, seq_num 12
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:12) to host localhost
++ volume match: have volume from index 12 to 12
-- vol match returns 1
13
-- svs: run 30, seq_num 13
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:13) to host localhost
++ volume match: have volume from index 13 to 13
-- vol match returns 1
14
-- svs: run 30, seq_num 14
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:14) to host localhost
++ volume match: have volume from index 14 to 14
-- vol match returns 1
15
-- svs: run 30, seq_num 15
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:15) to host localhost
++ volume match: have volume from index 15 to 15
-- vol match returns 1
16
-- svs: run 30, seq_num 16
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:16) to host localhost
++ volume match: have volume from index 16 to 16
-- vol match returns 1
17
-- svs: run 30, seq_num 17
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:17) to host localhost
++ volume match: have volume from index 17 to 17
-- vol match returns 1
18
-- svs: run 30, seq_num 18
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:18) to host localhost
++ volume match: have volume from index 18 to 18
-- vol match returns 1
19
-- svs: run 30, seq_num 19
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:19) to host localhost
++ volume match: have volume from index 19 to 19
-- vol match returns 1
20
-- svs: run 30, seq_num 20
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:20) to host localhost
++ volume match: have volume from index 20 to 20
-- vol match returns 1
21
-- svs: run 30, seq_num 21
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:21) to host localhost
++ volume match: have volume from index 21 to 21
-- vol match returns 1
22
-- svs: run 30, seq_num 22
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:22) to host localhost
++ volume match: have volume from index 22 to 22
-- vol match returns 1
23
-- svs: run 30, seq_num 23
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:23) to host localhost
++ volume match: have volume from index 23 to 23
-- vol match returns 1
24
-- svs: run 30, seq_num 24
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:24) to host localhost
++ volume match: have volume from index 24 to 24
-- vol match returns 1
25
-- svs: run 30, seq_num 25
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:25) to host localhost
++ volume match: have volume from index 25 to 25
-- vol match returns 1
26
-- svs: run 30, seq_num 26
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:26) to host localhost
++ volume match: have volume from index 26 to 26
-- vol match returns 1
27
-- svs: run 30, seq_num 27
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:27) to host localhost
++ volume match: have volume from index 27 to 27
-- vol match returns 1
28
-- svs: run 30, seq_num 28
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:28) to host localhost
++ volume match: have volume from index 28 to 28
-- vol match returns 1
29
-- svs: run 30, seq_num 29
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:29) to host localhost
++ volume match: have volume from index 29 to 29
-- vol match returns 1
30
-- svs: run 30, seq_num 30
completing orients from 'RLAPIS' to 'RLAPIS'
ART: sent images from volume (30:30) to host localhost
-- vol match returns 0
-- sorted fnames: from 30 to 30
-- RNI: read 0 new images (nerrs = 0)
-- fim_o: no sorting to do
-- read_image_files: 0 images, start = 30, proc = 0, tot = 30
ART: xim alloc: -1 -> 2568800 bytes
ART: ** failed to transmit EOR to afni @ localhost
- closing afni connection
- iochan error: iochan_goodcheck: no longer alive
ART: exit: closing afni control channel
++ writing dimon file list to dimon.files.run.030
++ oblique data: applying to3d -oblique_origin
set OutlierCheck =
set OutPrefix = OutBrick
to3d -assume_dicom_mosaic -prefix OutBrick_run_030 -time:zt 76 30 1.0sec alt+z -oblique_origin -@
++ to3d: AFNI version=AFNI_24.3.06 (Oct 31 2024) [64-bit]
++ Authored by: RW Cox
++ It is best to use to3d via the Dimon program.
++ DICOM WARNING: file 001_000030_000001_1.3.12.2.1107.5.2.61.237004.2024102815241190720020514.dcm has Rescale tags
setenv AFNI_DICOM_RESCALE YES to enforce them
++ Data detected to be oblique
++ DICOM WARNING: file 001_000030_000002_1.3.12.2.1107.5.2.61.237004.2024102815241266975020561.dcm has Rescale tags
setenv AFNI_DICOM_RESCALE YES to enforce them
++ DICOM WARNING: no more Rescale tags messages will be printed
*+ WARNING: Bad DICOM header - assuming oblique scaling direction!
*+ WARNING: *** ILLEGAL INPUTS (cannot save) ***
++ Counting images: total=2280 2D slices++ Each 2D slice is 130 X 130 pixels
++ Voxel dimensions: 1.6000 X 1.6000 X 1.6000 mm
++ Image data type = short
++ Reading images: ..........................................
++ Command line TR=1s ; Images TR=1s
++ Making widgets.....
final run statistics:
volume info :
slices : 1
z_first : -64.41
z_last : -64.41
z_delta : 1.6
oblique : yes
mos_nslices : 0
run # 30 : volumes = 30, first file (#0) = 001_000030_000001_1.3.12.2.1107.5.2.61.237004.2024102815241190720020514.dcm
++ Counting images: total=2280 2D slices
++ Each 2D slice is 130 X 130 pixels
++ Voxel dimensions: 1.6000 X 1.6000 X 1.6000 mm
++ Image data type = short
++ Reading images: ..........................................
++ Command line TR=1s ; Images TR=1s
++ Making widgets.....
final run statistics:
volume info :
slices : 1
z_first : -64.41
z_last : -64.41
z_delta : 1.6
oblique : yes
mos_nslices : 0
run # 30 : volumes = 30, first file (#0) = 001_000030_000001_1.3.12.2.1107.5.2.61.237004.2024102815241190720020514.dcm