AFNI version info (afni -ver): 24.2.01
Hi there,
I am using afni_proc.py to analyze my block design data. The data has already been preprocessed using fMRIPrep. I have two questions:
Question #1: Masks
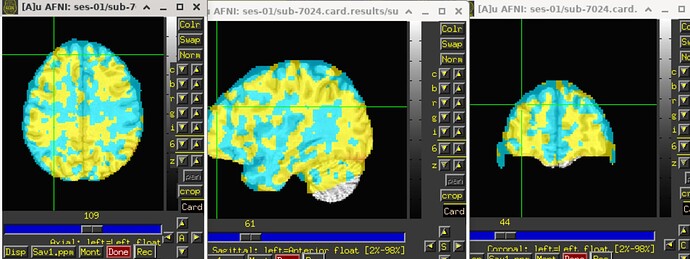
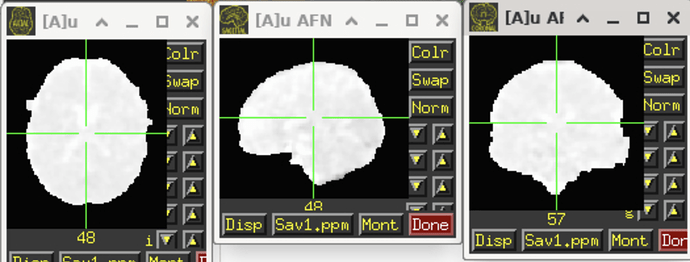
I decided to let afni create the mask using the epi data rather than load in the fMRIPrep mask. When I go to look at my subject-level data in the output, the mask looks wonky. There seems to be parts that extend beyond the cortex and don't fully match up with the anatomy. In some areas it almost seems like the mask has ears. Here are some images
Do you have any advice for this? Or what mask I should be using during the second level analysis? I don't want any results outside of the MNI template brain.
My second question pertains to the -regress_run_clustsim flag. I watched the AFNI bootcamp videos and it said that this flag applies cluster level thresholding at the individual level. I was wondering if this is necessary if I want to do cluster-level thresholding at the group level (p-FWE<.05) in addition to voxel thresholding (p<.001)? I don't want to overcorrect my data.
My script is pasted below.
Thank you as always for your help! This message board is fantastic ![]()
Meg
#!/bin/bash
# File containing the list of subjects
subject_file="/oscar/data/jbarredo/nstb/code/card/subjects_54.txt"
# Loop through each subject in the list
while read subID; do
# Set the session and task
sess='ses-01'
task='card'
afni_proc.py \
-subj_id $subID \
-out_dir /oscar/data/jbarredo/nstb/derivatives/afni/$subID/$sess/$subID.$task.results \
-copy_anat /oscar/data/jbarredo/nstb/derivatives/fmriprep/$subID/$sess/anat/${subID}_${sess}_space-MNI152NLin2009cAsym_desc-preproc_T1w.nii.gz \
-anat_has_skull no \
-dsets /oscar/data/jbarredo/nstb/derivatives/fmriprep/$subID/$sess/func/${subID}_${sess}_task-card_dir-AP_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz \
/oscar/data/jbarredo/nstb/derivatives/fmriprep/$subID/$sess/func/${subID}_${sess}_task-card_dir-PA_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz \
-blocks blur mask scale regress \
-tcat_remove_first_trs 0 \
-mask_apply epi \
-mask_epi_anat yes \
-blur_size 6.0 \
-regress_stim_times /oscar/data/jbarredo/nstb/derivatives/card/AFNI/${subID}_reward.1D \
/oscar/data/jbarredo/nstb/derivatives/card/AFNI/${subID}_punish.1D \
/oscar/data/jbarredo/nstb/derivatives/card/AFNI/${subID}_baseline.1D \
-regress_stim_labels reward punish baseline \
-regress_stim_types AM1 \
-regress_basis 'dmUBLOCK' \
-regress_polort 3 \
-regress_opts_3dD -jobs 4 \
-gltsym 'SYM: reward -baseline' \
-glt_label 1 reward_vs_baseline \
-gltsym 'SYM: punish -baseline' \
-glt_label 2 punish_vs_baseline \
-gltsym 'SYM: punish -reward' \
-glt_label 3 punish_vs_reward \
-gltsym 'SYM: reward -punish' \
-glt_label 4 reward_vs_punish \
-regress_motion_file /oscar/data/jbarredo/nstb/derivatives/card/motion/${subID}_motion_combined.txt \
-regress_est_blur_errts \
-regress_run_clustsim yes \
-html_review_style pythonic \
-execute
done < $subject_file