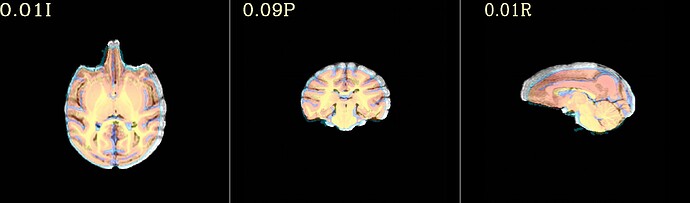
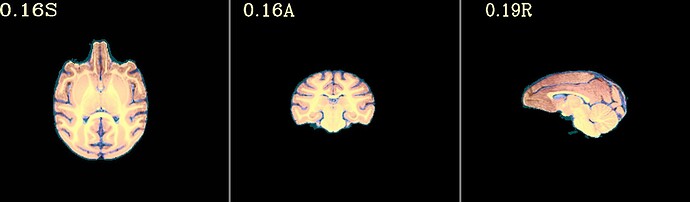
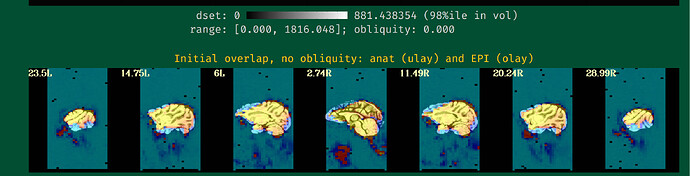
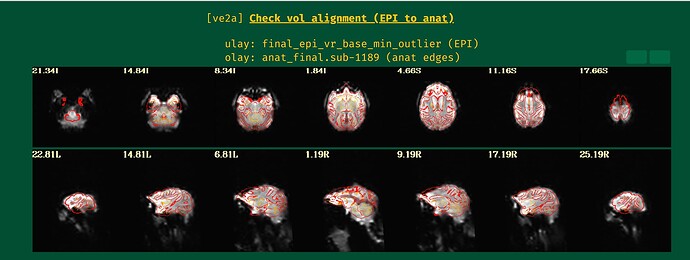
Hi all,
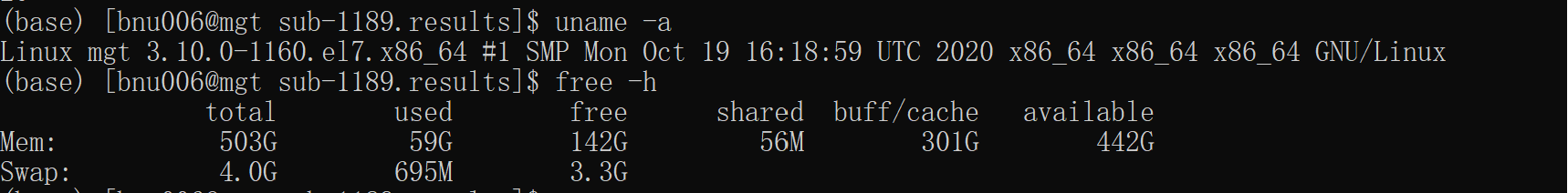
I have created my specific age T1 template firstly. Then I run Animan_warper to register T1 image to my template. Last, I run afni_proc.py. Unfortunately, afni_proc.py took about 14 hours for each subject. Is it normal and do I need to adjust my code to process rs-fmri data?The code I used is as follows:
set subj = $1
set ap_label = rsfmri_regressall
set dir_inroot = ${PWD:h}
set dir_log = ${dir_inroot}/logs
set dir_ref = ${dir_inroot}/data_age1_basic/template
set dir_basic = ${dir_inroot}/data_age1_basic
set dir_aw = ${dir_inroot}/data_13_aw/age1
set dir_ap = ${dir_inroot}/data_${ap_label}/age1
set sdir_basic = ${dir_basic}/${subj}
set sdir_anat = ${sdir_basic}/anat
set sdir_epi = ${sdir_basic}/func
set sdir_aw = ${dir_aw}/${subj}
set sdir_ap = ${dir_ap}/${subj}
set anat_orig = ${sdir_anat}/${subj}*T1w_bet.nii.gz
set anat_orig_ab = ${subj}_anat
set ref_base = ${dir_ref}/Template_Standard_Age1.nii
set ref_base_ab = template_age1
set dsets_epi = ( ${sdir_epi}/${subj}*task-rest_bold.nii.gz )
set anat_cp = ${sdir_aw}/${anat_orig_ab}_nsu.nii.gz
set dsets_NL_warp = ( ${sdir_aw}/${anat_orig_ab}_warp2std_nsu.nii.gz \
${sdir_aw}/${anat_orig_ab}_composite_linear_to_template.1D \
${sdir_aw}/${anat_orig_ab}_shft_WARP.nii.gz )
set nthr_avail = `afni_system_check.py -disp_num_cpu`
set nthr_using = `afni_check_omp`
echo "++ INFO: Using ${nthr_avail} of available ${nthr_using} threads"
setenv AFNI_COMPRESSOR GZIP
set ap_cmd = ${sdir_ap}/ap.cmd.${subj}
\mkdir -p ${sdir_ap}
cat <<EOF >! ${ap_cmd}
# -----------------------------------------------------------------
setenv OMP_NUM_THREADS 10
afni_proc.py \
-subj_id ${subj} \
-blocks tshift align tlrc volreg blur mask scale regress \
-dsets ${dsets_epi} \
-copy_anat ${anat_cp} \
-anat_has_skull no \
-anat_uniform_method none \
-radial_correlate_blocks tcat volreg regress \
-radial_correlate_opts -sphere_rad 14 \
-tcat_remove_first_trs 10 \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-volreg_warp_dxyz 0.5 \
-volreg_compute_tsnr yes \
-align_opts_aea -cost nmi \
-check_flip -feature_size 0.5 \
-align_unifize_epi local \
-tlrc_base ${ref_base} \
-tlrc_NL_warp \
-tlrc_NL_warped_dsets ${dsets_NL_warp} \
-blur_size 2.5 \
-mask_segment_anat yes \
-mask_segment_erode yes \
-mask_import Tvent /data/home/bnu006/UW-Madison_Rhesus_MRI/preprocess/data_age1_basic/template/ventricles_in_template_age1.nii \
-mask_intersect Svent CSF Tvent \
-regress_ROI WMe Svent \
-regress_ROI_per_run WMe Svent \
-regress_motion_per_run \
-regress_apply_mot_types demean deriv \
-regress_polort 2 \
-regress_bandpass 0.01 0.1 \
-regress_est_blur_errts \
-regress_est_blur_epits \
-regress_run_clustsim no \
-html_review_style pythonic
EOF
cd ${sdir_ap}
tcsh -xef ${ap_cmd} |& tee output.ap.cmd.${subj}
time tcsh -xef proc.${subj} |& tee output.proc.${subj}
echo "++ FINISHED AP: ${ap_label}"
exit 0