Hello!
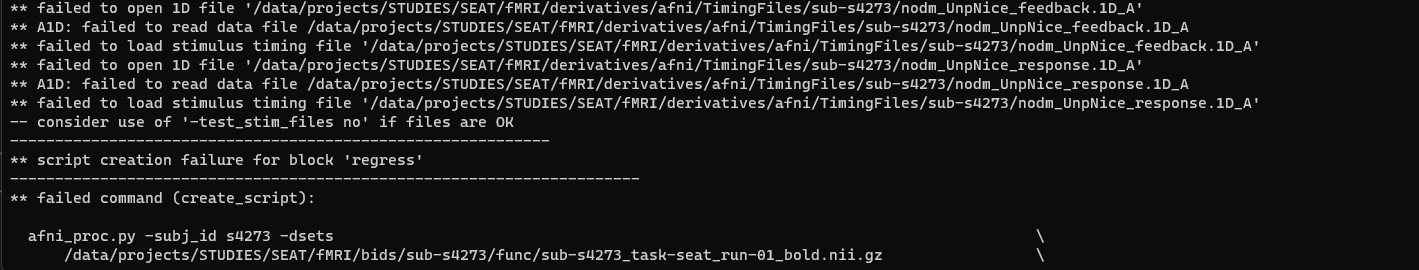
I am trying to rerun some individual level analyses and am running into 2 errors that I cannot understand:
I have checked the integrity of the divorced timing files (onset only) with file_tool -test -infile xxx but the output declares no bad characters and file type: Unix. I even created the FIXED.1D file with the -prefix command and the output was identical with no discernable changes from the original 1D timing file.
I don't know what to make of the failure to create script error, as several other folks seemed to have this problem fixed by fixing the timing files or adding a command which we already use in this code. Here is the code:
# **CHANGE ME**: Specify subject numbers in a single row. Include the s (e.g., s305)
set subjects = ( s4273 s305 s1571 s1712 s1146 s1585 s1603 s1622 s1671 s1696 s1718 s1754 s4075 s4096 s4148 s4142 s318 s563 s651 s747 s1541 s1562 s1573 s1577 s1587 s1591 s1610 s1614 s1629 s1639 s1692 s1693 s1714 s1722 s1746 s4084 s4089 s4130 s4136 s1521 s4111 s4128 s4164 s4201 s4207 s4209 s4211 s4220 s4242 s4262 s4228 )
#run s4273 s305 s1571 s1712 s1146 s1585 s1603 s1622 s1671 s1696 s1718 s1754 s4075 s4096 s4148 s4142 s318 s563 s651 s747 s1541 s1562 s1573 s1577 s1587
#s1591 s1610 s1614 s1629 s1639 s1692 s1693 s1714 s1722 s1746 s4084 s4089 s4130 s4136 s1521 s4111 s4128 s4164 s4201 s4207 s4209 s4211 s4220 s4242 s4262
#s4228
######################################################
# SPECIFY ANALYSIS NAME, MOTION AND TR EXCLUSION PREFERENCES
######################################################
# **CHECK ME**: Provide the name of the GLM you are running; The motion censor value will be appended to the end
set GLM = SEATReputationBLOCK
# **CHECK ME**: Specify the maximum amount of motion allowed without censoring
set motion_max = 1
######################################################
# SPECIFY LOCATIONS
######################################################
# Specify the top-most directory associated with the study
set topdir = /data/projects/STUDIES/SEAT/fMRI
# Specify the directory where subjects' neuroimaging data that have been run through the BIDS conversion
set subjfmridata = /data/projects/STUDIES/SEAT/fMRI/bids
# Specify the directory where subjects' timing/behavioral data are located
set subjecttiming = /data/projects/STUDIES/SEAT/fMRI/derivatives/afni/TimingFiles
# Specify the directory where individual subject analysis afni_proc.py and results will be written
set results = /data/projects/STUDIES/SEAT/fMRI/derivatives/afni/IndvlLvlAnalyses
set anat_dir = /data/projects/STUDIES/SEAT/fMRI/derivatives/afni/ssw
############################################################################################
# THE ANALYSIS SCRIPT BEGINS HERE
############################################################################################
######################################################
# NAVIGATE TO PROPER LOCATION, AND FINISH SET-UP
######################################################
# Go to the results tree and work from there
cd $results
# For each of the subjects, specified in the subjects list...
foreach subj ( $subjects )
# Make a new subject results folder
mkdir $subj
# Change directories into that folder to work
cd $subj
##### Set subject-specific data to be used in afni_proc ####
# Specify the subject's fMRI data directory path
set subj_dir = $subjfmridata/sub-$subj
# Specify the folder where the unmarried (onset only) stimulus timing files are located
set stimdurmoddir = $subjecttiming/sub-$subj
######################################################
# SPECIFY PARAMETERS FOR AFNI_PROC.PY
######################################################
afni_proc.py -subj_id $subj \
-dsets $subj_dir/func/sub-${subj}_task-seat_run-01_bold.nii.gz \
$subj_dir/func/sub-${subj}_task-seat_run-02_bold.nii.gz \
$subj_dir/func/sub-${subj}_task-seat_run-03_bold.nii.gz \
-scr_overwrite \
-script $results/$subj/proc.$subj.$GLM.${motion_max}mm \
-out_dir $subj.results.$GLM.${motion_max}mm \
-blocks despike tshift align tlrc volreg blur mask scale regress \
-copy_anat $anat_dir/sub-${subj}/anatSS.$subj.nii \
-anat_has_skull no \
-anat_follower anat_w_skull anat $anat_dir/sub-${subj}/anatU.$subj.nii \
-mask_epi_anat yes \
-tlrc_base MNI152_2009_template_SSW.nii.gz \
-tshift_align_to -tzero 0 \
-align_opts_aea \
-giant_move \
-cost lpc+ZZ \
-AddEdge \
-anat_uniform_method unifize \
-tlrc_NL_warp \
-tlrc_NL_warped_dsets \
$anat_dir/sub-${subj}/anatQQ.${subj}.nii \
$anat_dir/sub-${subj}/anatQQ.${subj}.aff12.1D \
$anat_dir/sub-${subj}/anatQQ.${subj}_WARP.nii \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-volreg_warp_dxyz 3 \
-blur_size 6 \
-mask_dilate 1 \
-scale_max_val 200 \
-regress_censor_outliers 0.1 \
-regress_motion_per_run \
-regress_censor_motion $motion_max \
-regress_est_blur_epits \
-regress_est_blur_errts \
-regress_run_clustsim yes \
-regress_compute_fitts \
-html_review_style pythonic \
-regress_stim_times \
$stimdurmoddir/nodm_Mean_anticipation.1D_A \
$stimdurmoddir/nodm_Mean_feedback.1D_A \
$stimdurmoddir/nodm_Mean_response.1D_A \
$stimdurmoddir/nodm_Nice_anticipation.1D_A \
$stimdurmoddir/nodm_Nice_feedback.1D_A \
$stimdurmoddir/nodm_Nice_response.1D_A \
$stimdurmoddir/nodm_Unp_anticipation.1D_A \
$stimdurmoddir/nodm_UnpMean_feedback.1D_A \
$stimdurmoddir/nodm_UnpMean_response.1D_A \
$stimdurmoddir/nodm_UnpNice_feedback.1D_A \
$stimdurmoddir/nodm_UnpNice_response.1D_A \
-regress_stim_labels \
Ant.Mean \
FB.Mean \
Resp.Mean \
Ant.Nice \
FB.Nice \
Resp.Nice \
Ant.Unp \
FB.UnpMean \
Resp.UnpMean \
FB.UnpNice \
Resp.UnpNice \
-regress_stim_types \
'times' \
-regress_basis_multi \
'BLOCK(3,1)' \
'BLOCK(2,1)' \
'BLOCK(3,1)' \
'BLOCK(3,1)' \
'BLOCK(2,1)' \
'BLOCK(3,1)' \
'BLOCK(3,1)' \
'BLOCK(2,1)' \
'BLOCK(3,1)' \
'BLOCK(2,1)' \
'BLOCK(3,1)' \
-regress_make_ideal_sum IDEAL_sum.1D \
-regress_opts_3dD \
-GOFORIT 10 \
-allzero_OK \
-num_glt 15 \
-gltsym 'SYM: +Ant.Mean +FB.Mean +Resp.Mean +Ant.Nice +FB.Nice +Resp.Nice +Ant.Unp +FB.UnpMean +Resp.UnpMean +FB.UnpNice +Resp.Nice' -glt_label 1 Task.V.BL \
-gltsym 'SYM: +Ant.Mean +Ant.Nice +Ant.Unp' -glt_label 2 Ant.V.BL \
-gltsym 'SYM: +Ant.Mean -Ant.Nice' -glt_label 3 Ant.Mean.V.Nice \
-gltsym 'SYM: +Ant.Mean -Ant.Unp' -glt_label 4 Ant.Mean.V.Unp \
-gltsym 'SYM: +Ant.Nice -Ant.Unp' -glt_label 5 Ant.Nice.V.Unp \
-gltsym 'SYM: +FB.Mean +FB.Nice +FB.UnpMean +FB.UnpNice' -glt_label 6 FB.V.BL \
-gltsym 'SYM: +FB.Mean +FB.UnpMean -FB.Nice -FB.UnpNice' -glt_label 7 FB.Mean.V.Nice \
-gltsym 'SYM: +FB.Mean -FB.UnpMean' -glt_label 8 FB.Mean.V.UnpMean \
-gltsym 'SYM: +FB.Nice -FB.UnpNice' -glt_label 9 FB.Nice.V.UnpNice \
-gltsym 'SYM: +FB.Mean +FB.Nice -FB.UnpMean -FB.UnpNice' -glt_label 10 FB.Pred.V.Unp \
-gltsym 'SYM: +Resp.Mean +Resp.Nice +Resp.UnpMean +Resp.UnpNice' -glt_label 11 Resp.V.BL \
-gltsym 'SYM: +Resp.Mean -Resp.Nice' -glt_label 12 Resp.Mean.V.Nice \
-gltsym 'SYM: +Resp.Mean -Resp.UnpMean' -glt_label 13 Resp.Mean.V.UnpMean \
-gltsym 'SYM: +Resp.Nice -Resp.UnpNice' -glt_label 14 Resp.Nice.V.UnpNice \
-gltsym 'SYM: +Resp.Mean +Resp.Nice -Resp.UnpMean -Resp.UnpNice' -glt_label 15 Resp.Pred.V.Unp \
-cbucket cbucket.stats.$subj \
-jobs 30 \
cd ..
end