AFNI version info (afni -ver): Version AFNI_24.3.05 'Elagabalus
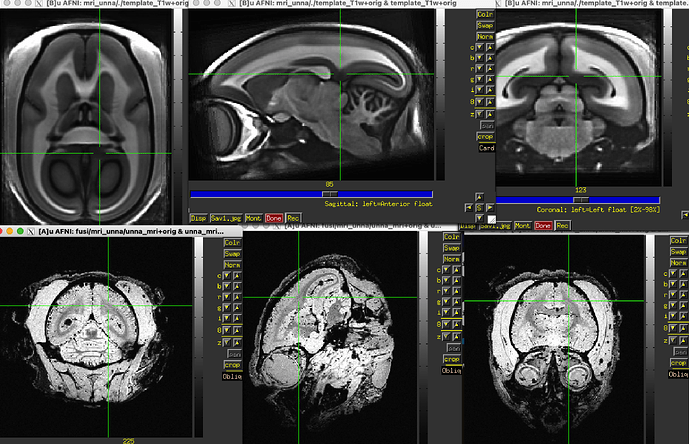
Hiii, I have a mamorset MRI (carcass) trying to align to a T1 template. It shouldn't be a problem with animal_warper, but the result looks weird/distorted.
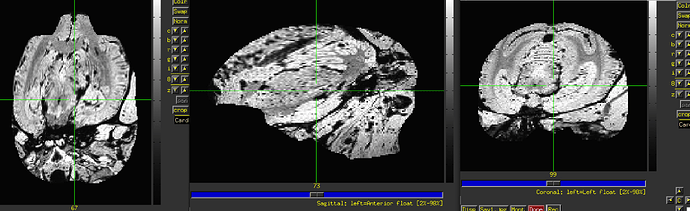
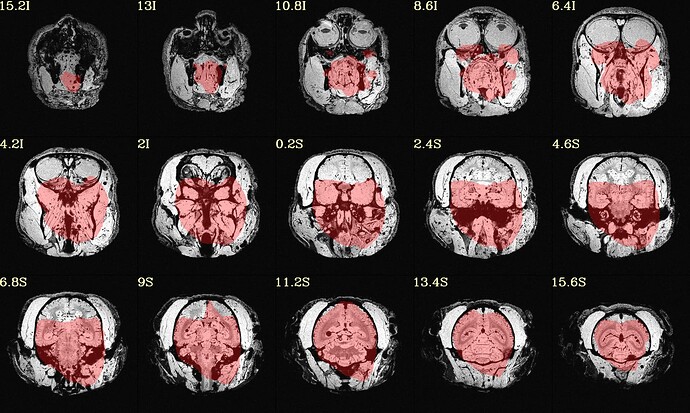
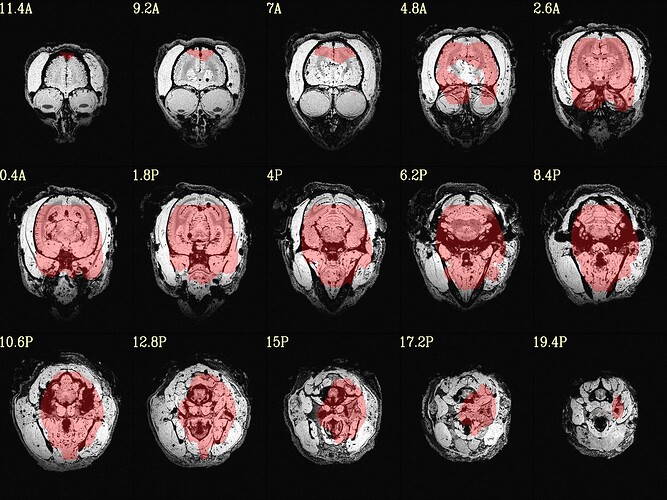
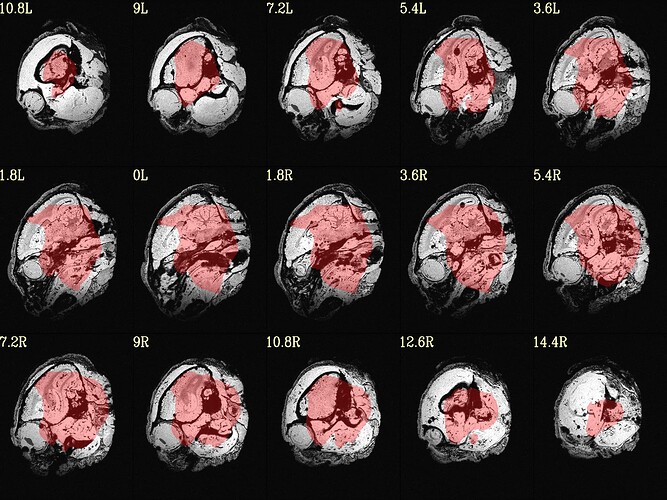
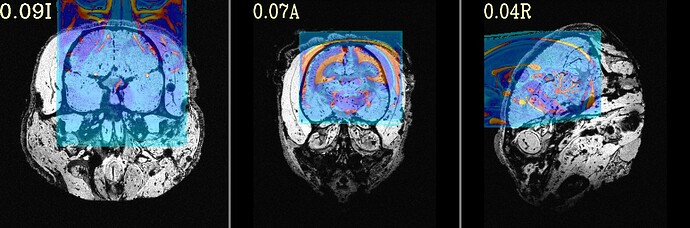
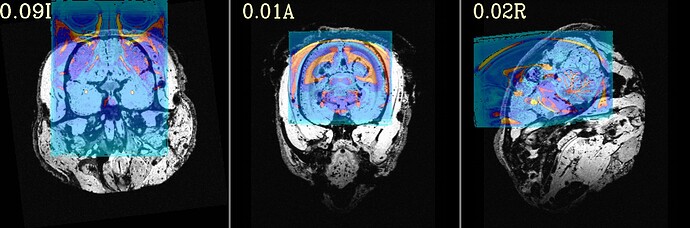
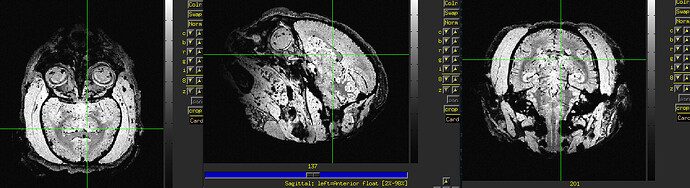
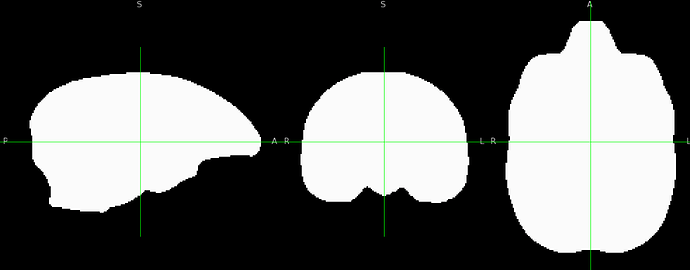
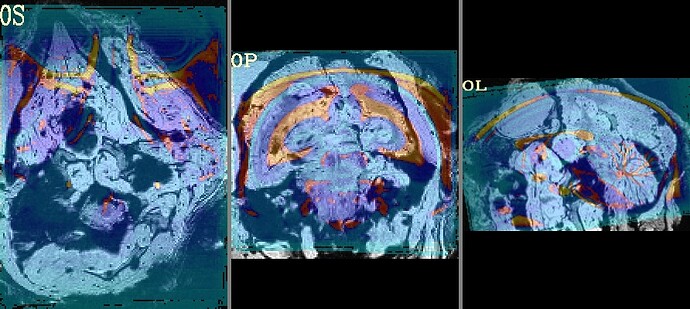
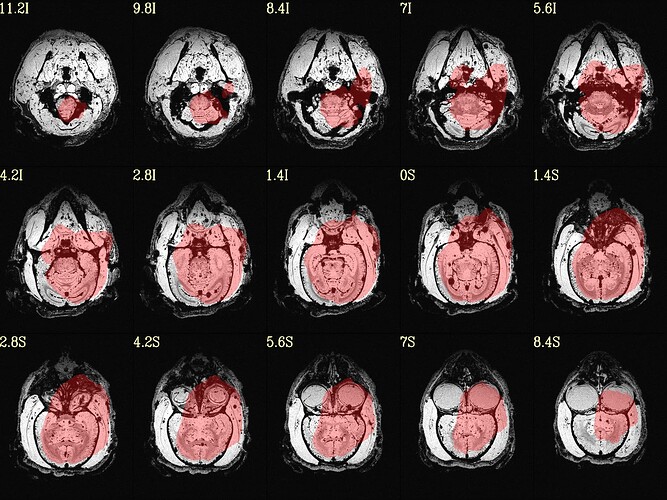
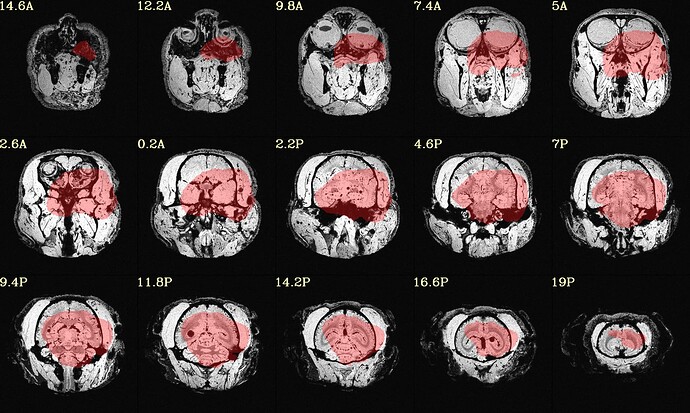
The final QC images don't look very good. (qc_02 I assume those are the final qc?)
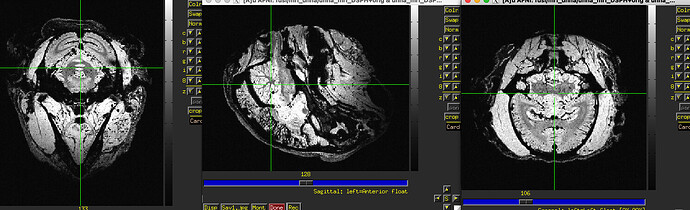
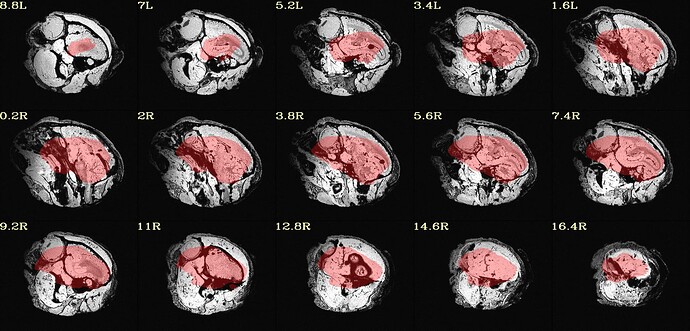
as you can see, the sagittal part is a bit weird. It looks like there is an oblique (checked using 3dinfo -is_oblique got 1). but then I ran adjunct_deob_around_origin, the image remain the same.
here the first row is the template, the second raw is the carcass MRI. Maybe it's not an oblique problem? what would be the problem then? there is not sphinix problem either.
Thank you!
# Check if input file is oblique using 3dinfo
is_oblique=0
if [ "$(3dinfo -is_oblique "$input_file")" != "0" ]; then
is_oblique=1
fi
if [ "$is_oblique" -eq 1 ]; then
echo "Input file is oblique. Using adjunct_deob_around_origin to handle obliquity." >> "${stdout_log}"
base_filename=$(strip_extension "$original_input_file")
temp_prefix="${base_filename}_deob"
echo "Base filename: $base_filename" >> "${stdout_log}"
echo "Temp prefix: $temp_prefix" >> "${stdout_log}"
# Run adjunct_deob_around_origin
echo "Running adjunct_deob_around_origin..." >> "${stdout_log}"
if ! adjunct_deob_around_origin -input "${original_input_file}" -prefix "${preproc_dir}/${temp_prefix}" >> "${stdout_log}" 2>> "${stderr_log}"; then
echo "ERROR: adjunct_deob_around_origin failed" >> "${stderr_log}"
exit 1
fi
# List files in output directory for debugging
echo "Files in preprocessing directory:" >> "${stdout_log}"
ls -l "${preproc_dir}" >> "${stdout_log}"
# Check for AFNI format first (most likely output)
if [ -f "${preproc_dir}/${temp_prefix}+orig.HEAD" ]; then
echo "Found AFNI format file, converting to NIFTI..." >> "${stdout_log}"
# Convert AFNI to NIFTI
if ! 3dAFNItoNIFTI -prefix "${preproc_dir}/${temp_prefix}" "${preproc_dir}/${temp_prefix}+orig" >> "${stdout_log}" 2>> "${stderr_log}"; then
echo "ERROR: Failed to convert AFNI to NIFTI" >> "${stderr_log}"
exit 1
fi
# Wait for file to be created
sleep 1
fi
# Now check for NIFTI files
if [ -f "${preproc_dir}/${temp_prefix}.nii.gz" ]; then
input_file="${preproc_dir}/${temp_prefix}.nii.gz"
elif [ -f "${preproc_dir}/${temp_prefix}.nii" ]; then
input_file="${preproc_dir}/${temp_prefix}.nii"
else
echo "ERROR: Could not find output file. Checked for:" >> "${stderr_log}"
echo " - ${preproc_dir}/${temp_prefix}.nii.gz" >> "${stderr_log}"
echo " - ${preproc_dir}/${temp_prefix}.nii" >> "${stderr_log}"
echo " - ${preproc_dir}/${temp_prefix}+orig.HEAD" >> "${stderr_log}"
echo "Directory contents:" >> "${stderr_log}"
ls -l "${preproc_dir}" >> "${stderr_log}"
exit 1
fi
echo "Using deobliqued file: $input_file" >> "${stdout_log}"
else
echo "Input file is not oblique. No deobliquing needed." >> "${stdout_log}"
input_file="$original_input_file"
fi
animal_warper_cmd="@animal_warper -input \"$input_file\" -base \"$base_file\" -outdir \"$output_dir\" -skullstrip \"$skullstrip_file\" -ok_to_exist"
# Debug: Print the constructed command
echo "Running @animal_warper with the following command:" >> "${stdout_log}"
echo "$animal_warper_cmd" >> "${stdout_log}"
# Run the command using eval
eval "$animal_warper_cmd" >> "${stdout_log}" 2>> "${stderr_log}"