AFNI version info (afni -ver): Precompiled binary linux_centos_7_64: Jun 21 2023 (Version AFNI_23.1.08 'Publius Helvius Pertinax')
Hello AFNI gurus!
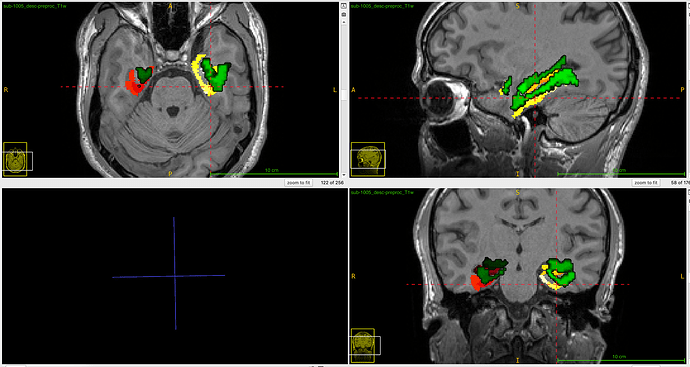
I'm trying to resample a manually-segmented hippocampal mask to functional space to extract beta values from it. But for some reason, 3dresample seems to shift the dataset a lot. In the attached image, the reddish dataset is the original ROI, and the green is the ROI after resampling.
This is resample command (nothing fancy!):
3dresample -input sub-1004_space-T1w_desc-HandSeg_mask.nii.gz \
-master sub-1004_task-aim_run-01_space-T1w_desc-preproc_bold.nii.gz \
-prefix sub-1004_space-T1w_desc-HandSeg_mask_resampled.nii.gz
The functional and T1w data was preprocessed using fmriprep (v23.0.2). The hippocampal ROI was created by manually segmenting the hippocampus on the individual's T2w scan. The T2 was then aligned to the preprocessed T1 using ANTs. And this transformation matrix was applied to the ROI to bring it to T1 space.
Could this be happening because, somehow, fmriprep puts the T1 and functional data in different spaces, views, etc?
Here are the outputs from 3dinfo on these datasets:
Preprocessed T1:
Identifier Code: NII_lq875bLvWFd_7NgQ7XaCiA Creation Date: Mon Sep 16 20:20:51 2024
Template Space: ORIG
Dataset Type: Anat Bucket (-abuc)
Byte Order: LSB_FIRST {assumed} [this CPU native = LSB_FIRST]
Storage Mode: NIFTI
Storage Space: 46,137,344 (46 million) bytes
Geometry String: "MATRIX(-0.999367,-0.018268,-0.030602,94.54953,0.018787,-0.999683,-0.016754,105.0525,-0.030287,-0.017319,0.999391,-122.564):176,256,256"
Data Axes Tilt: Oblique (2.042 deg. from plumb)
Data Axes Approximate Orientation:
first (x) = Left-to-Right
second (y) = Posterior-to-Anterior
third (z) = Inferior-to-Superior [-orient LPI]
R-to-L extent: -80.451 [R] -to- 94.550 [L] -step- 1.000 mm [176 voxels]
A-to-P extent: -149.947 [A] -to- 105.052 [P] -step- 1.000 mm [256 voxels]
I-to-S extent: -122.564 [I] -to- 132.436 [S] -step- 1.000 mm [256 voxels]
Number of values stored at each pixel = 1
-- At sub-brick #0 '?' datum type is float
Preprocessed functional:
Identifier Code: NII_FfAmt0ywfFlY1QiUGQBs7Q Creation Date: Mon Sep 16 20:21:42 2024
Template Space: TLRC
Dataset Type: Echo Planar (-epan)
Byte Order: LSB_FIRST {assumed} [this CPU native = LSB_FIRST]
Storage Mode: NIFTI
Storage Space: 557,062,800 (557 million) bytes
Geometry String: "MATRIX(-1.705,0,0,72.38453,0,-1.705,0,75.94521,0,0,1.7,-43.88039):85,105,83"
Data Axes Tilt: Plumb
Data Axes Orientation:
first (x) = Left-to-Right
second (y) = Posterior-to-Anterior
third (z) = Inferior-to-Superior [-orient LPI]
R-to-L extent: -70.835 [R] -to- 72.385 [L] -step- 1.705 mm [ 85 voxels]
A-to-P extent: -101.375 [A] -to- 75.945 [P] -step- 1.705 mm [105 voxels]
I-to-S extent: -43.880 [I] -to- 95.520 [S] -step- 1.700 mm [ 83 voxels]
Number of time steps = 188 Time step = 1.87000s Origin = 0.00000s
-- At sub-brick #0 '?' datum type is float
-- At sub-brick #1 '?' datum type is float
-- At sub-brick #2 '?' datum type is float
** For info on all 188 sub-bricks, use '3dinfo -verb' **
Manually segmented hippocampal ROI, aligned to T1 using ANTs:
Dataset File: sub-1005_space-T1w_desc-HandSeg_mask.nii.gz
Identifier Code: NII_AaC_TgbYphvAalPxsUF1Kw Creation Date: Mon Sep 16 20:22:11 2024
Template Space: ORIG
Dataset Type: Anat Bucket (-abuc)
Byte Order: LSB_FIRST {assumed} [this CPU native = LSB_FIRST]
Storage Mode: NIFTI
Storage Space: 46,137,344 (46 million) bytes
Geometry String: "MATRIX(-0.999367,-0.018268,-0.030602,94.54953,0.018787,-0.999683,-0.016754,105.0525,-0.030287,-0.017319,0.999391,-122.564):176,256,256"
Data Axes Tilt: Oblique (2.042 deg. from plumb)
Data Axes Approximate Orientation:
first (x) = Left-to-Right
second (y) = Posterior-to-Anterior
third (z) = Inferior-to-Superior [-orient LPI]
R-to-L extent: -80.451 [R] -to- 94.550 [L] -step- 1.000 mm [176 voxels]
A-to-P extent: -149.948 [A] -to- 105.052 [P] -step- 1.000 mm [256 voxels]
I-to-S extent: -122.564 [I] -to- 132.436 [S] -step- 1.000 mm [256 voxels]
Number of values stored at each pixel = 1
-- At sub-brick #0 '?' datum type is float
Finally, here is the comparison of the 3 datasets:
3dinfo -same_all_grid sub-1005_desc-preproc_T1w.nii.gz sub-1005_task-aim_run-01_space-T1w_desc-preproc_bold.nii.gz sub-1005_space-T1w_desc-HandSeg_mask.nii.gz
0 0 1 0 0
0 0 1 0 0
1 1 1 1 1
Strangely, the same thing happens even if I resample the ROI by explicitly setting -dxyz instead of providing a master dataset. So it may not even be the space/view issue?
Thank you, and sorry for the lengthy message!!
Mrinmayi