Dear AFNI experts,
I am using 3dLMEr to analyze my data at the group level as well as FWE correction.
The experiment is a within-subjects design with two factors (type: two levels -- ME, MI; task: three levels -- short, medium, long).
I want to make sure that the 3dLMEr script is correct? Alhough there are no errors in the terminal after running.The code is as follows:
AFNI version info (afni -ver): 23.1.00
3dLMEr -prefix group_result/LMEr -jobs 4 \
-mask mask \
-model 'type*task+(1|Subj)+(1|Subj:type)+(1|Subj:task)' \
-SS_type 3 \
-gltCode ME 'type : 1*ME' \
-gltCode MI 'type : 1*MI' \
-gltCode short 'task : 1*short' \
-gltCode middle 'task : 1*middle' \
-gltCode long 'task : 1*long' \
-gltCode ME-MI 'type : 1*ME -1*MI' \
-gltCode short-middle 'task : 1*short -1*middle' \
-gltCode short-long 'task : 1*short -1*long' \
-gltCode middle-long 'task : 1*middle -1*long' \
-glfCode type_task 'type : 1*ME -1*MI task : 1*short -1*middle & 1*short -1*long' \
-glfCode ME_task 'type : 1*ME task : 1*short -1*middle & 1*short -1*long' \
-glfCode MI_task 'type : 1*MI task : 1*short -1*middle & 1*short -1*long' \
-glfCode type_short 'type : 1*ME -1*MI task : 1*short' \
-glfCode type_middle 'type : 1*ME -1*MI task : 1*middle' \
-glfCode type_long 'type : 1*ME -1*MI task : 1*long' \
-dataTable @dataTable.txt
Using 3dinfo -verb to view the LMEr+tlrc file, there are 27 sub-bricks in total:
sub-brick #21 for the interaction between the type and task;
sub-brick #22/23 for the main effect of task at the ME/MI type;
sub-brick #24/25/26 for the main effect of type at the short/middle/long task;
What do the first three sub-bricks mean?
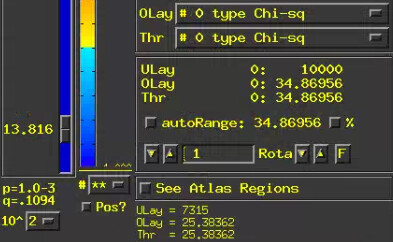
-- At sub-brick #0 'type Chi-sq' datum type is float: 0 to 34.8696
statcode = fict; statpar = 2
-- At sub-brick #1 'task Chi-sq' datum type is float: 0 to 36.1903
statcode = fict; statpar = 2
-- At sub-brick #2 'type:task Chi-sq' datum type is float: 0 to 22.8851
statcode = fict; statpar = 2
-- At sub-brick #3 'ME' datum type is float: -3.69596 to 4.00132
-- At sub-brick #4 'ME Z' datum type is float: -25.1851 to 31.6987
statcode = fizt
...
-- At sub-brick #19 'middle-long' datum type is float: -2.84857 to 1.23572
-- At sub-brick #20 'middle-long Z' datum type is float: -9.37283 to 3.76961
statcode = fizt
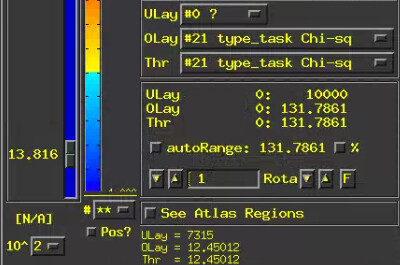
-- At sub-brick #21 'type_task Chi-sq' datum type is float: 0 to 131.786
-- At sub-brick #22 'ME_task Chi-sq' datum type is float: 0 to 101.148
-- At sub-brick #23 'MI_task Chi-sq' datum type is float: 0 to 114.847
-- At sub-brick #24 'type_short Chi-sq' datum type is float: 0 to 52.009
-- At sub-brick #25 'type_middle Chi-sq' datum type is float: 0 to 72.9838
-- At sub-brick #26 'type_long Chi-sq' datum type is float: 0 to 72.203
This is my first time using 3dLMEr and I don't know how to view the Chi-qs results.For Z-score, set the overlay sub-brick(e.g. sub-brick #3 'ME') and the threshold sub-brick(e.g. sub-brick #4 'ME Z'), then check the surviving voxel size by adjusting the p-value.
How to view the Chi-qs results? Set the overlay sub-brick same with the threshold sub-brick(e.g. sub-brick #0 'type Chi-sq')? I tried to look at the sub-brick #0 and the sub-brick #21. I've found that the sub-brick #0 can set a p-value, but the sub-brick #21 shows a p-value of [N/A].Why is this happening?
Please advise on FWE correction.
For general linear regression at the individual level, an errts file was generated for each participant. Average individual level errts file and perform 3dFWHMx -mask mask -input errts.mean_all_subj_REML+tlrc -acf. Is it right to do so? I see that 3dLMEr has a -resid option, can I use this residual to calculate directly? Which way is more reasonable?
I see that Gang mentioned a soft cluster threshold way, it doesn't make adjustments to the results?Alternatively, you may simply adopt a soft cluster threshold (e.g., 20 voxels at the p-value of 0.01), and use a highlight-but-not-hide approach as suggested in this recent paper.
Thanks,
Yang