AFNI version info (afni -ver):
Precompiled binary linux_ubuntu_16_64: Feb 22 2024 (Version AFNI_24.0.08 'Caracalla')
Very strange behavior encountered with trying to run 3dLMEr.
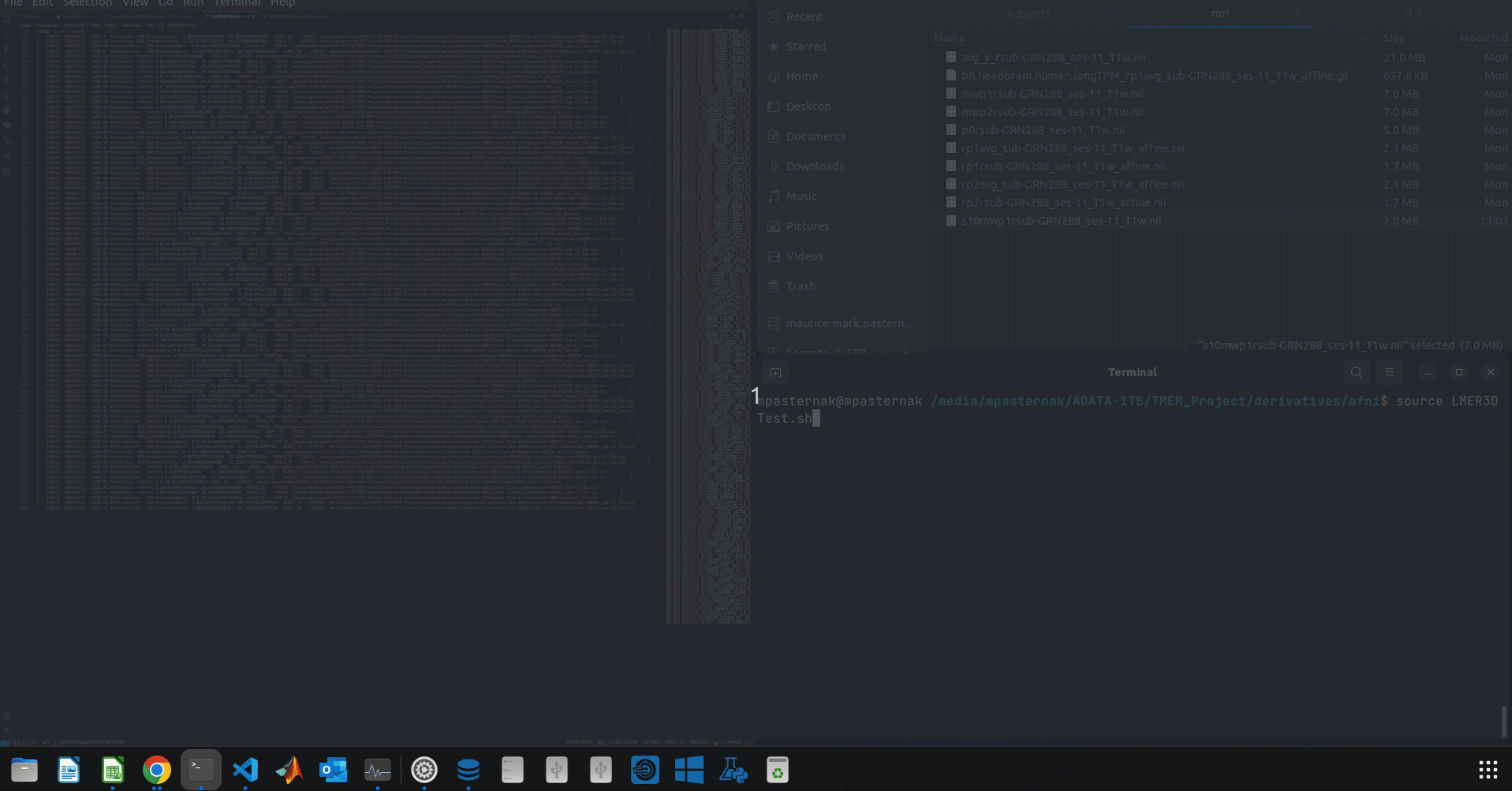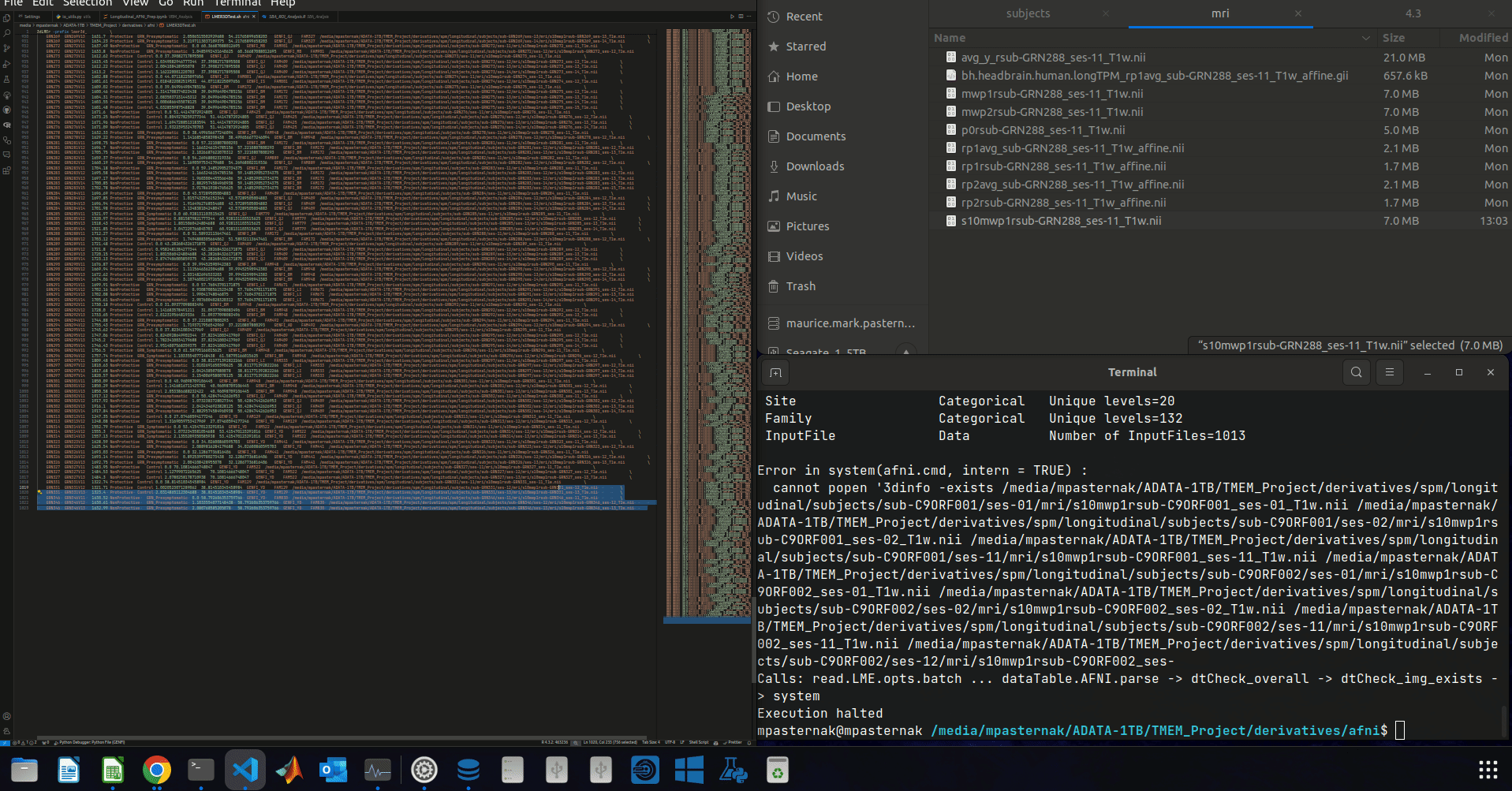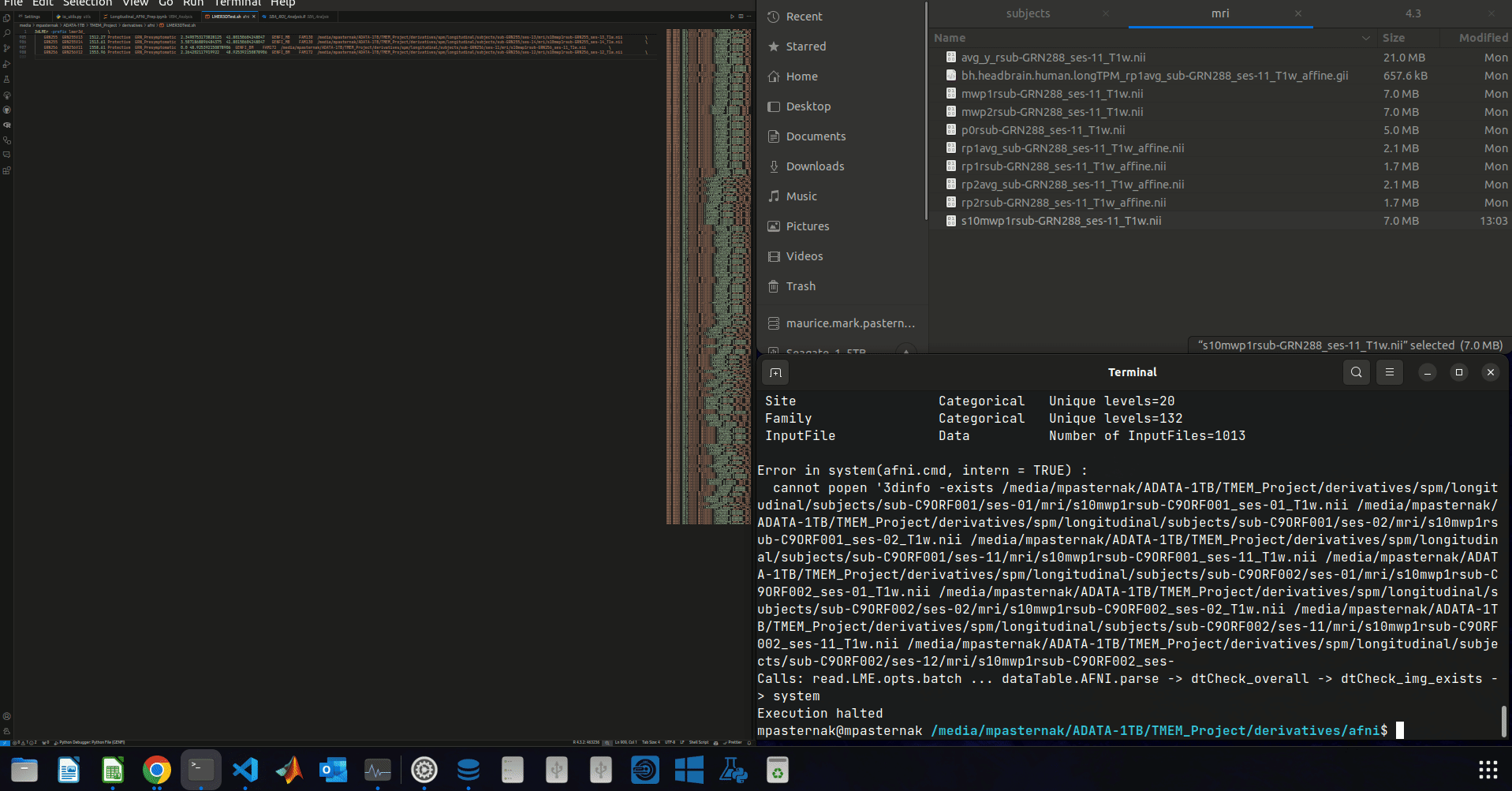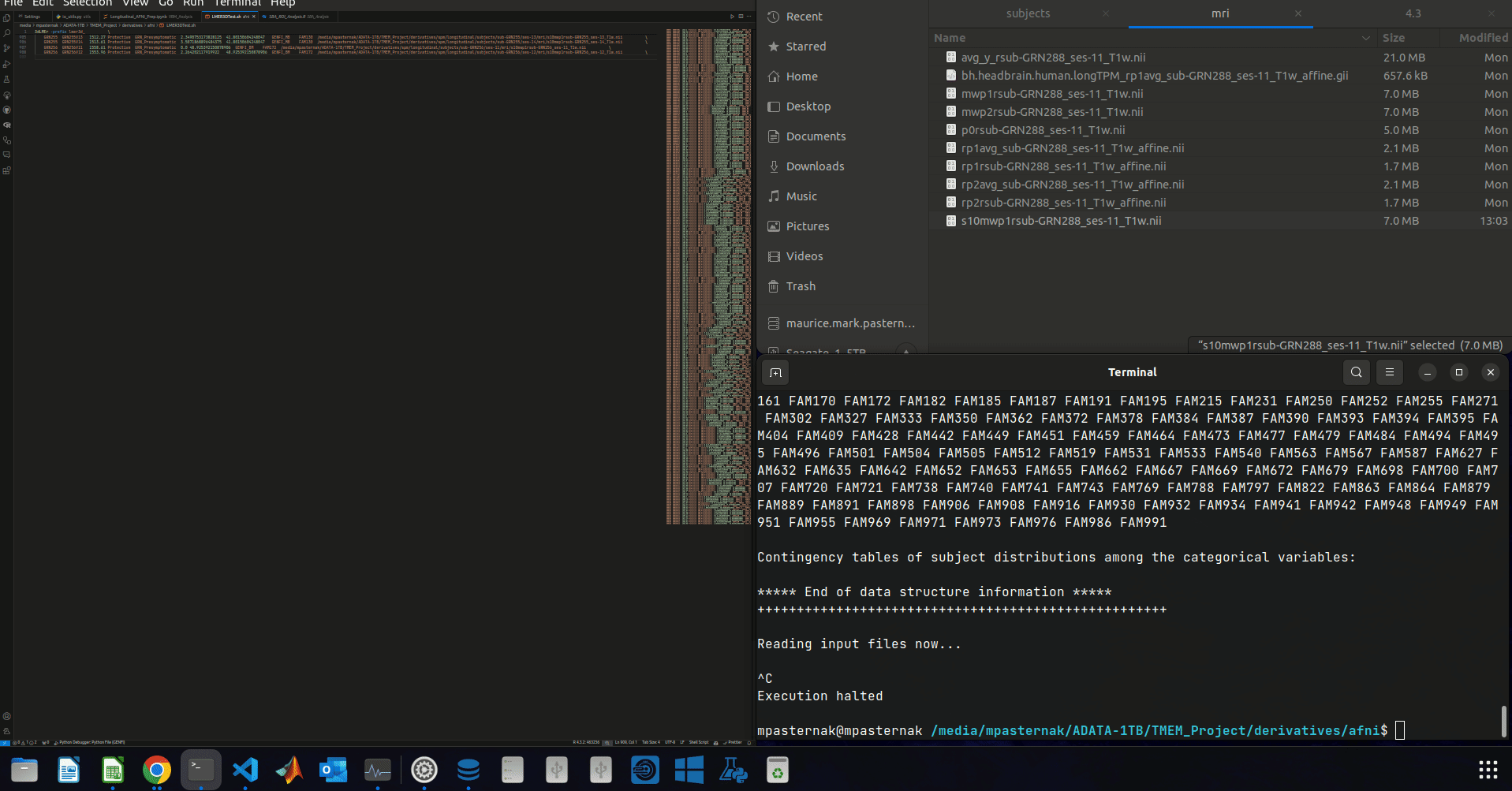
^ Let me just clarify what's going on in the gif:
- I try to run the 3dLMEr script. It fails.
- I delete a few rows from the data table. The script then runs.
And for the record, this seems to be independent of which rows from the data table I delete. As long as enough rows are deleted.
I prepared a 3dLMEr script programatically (you have likely heard of Pydantic for validation) and have validated it meets all 3dLMEr criteria (i.e. Subj column first, InputPath last, all files exist, etc. etc.). So I know the script isn't the issue here. Curiously, the script runs fine when I remove a large chunk of my subjects. But as soon as the number of subjects passes some critical number, afni experiences this issue:
Error in system(afni.cmd, intern = TRUE) :
cannot popen '3dinfo -exists /media/mpasternak/ADATA-1TB/TMEM_Project/derivatives/spm/longitudinal/subjects/sub-C9ORF001/ses-01/mri/s10mwp1rsub-C9ORF001_ses-01_T1w.nii /media/mpasternak/ADATA-1TB/TMEM_Project/derivatives/spm/longitudinal/subjects/sub-C9ORF001/ses-02/mri/s10mwp1rsub-C9ORF001_ses-02_T1w.nii /media/mpasternak/ADATA-1TB/TMEM_Project/derivatives/spm/longitudinal/subjects/sub-C9ORF001/ses-11/mri/s10mwp1rsub-C9ORF001_ses-11_T1w.nii /media/mpasternak/ADATA-1TB/TMEM_Project/derivatives/spm/longitudinal/subjects/sub-C9ORF002/ses-01/mri/s10mwp1rsub-C9ORF002_ses-01_T1w.nii /media/mpasternak/ADATA-1TB/TMEM_Project/derivatives/spm/longitudinal/subjects/sub-C9ORF002/ses-02/mri/s10mwp1rsub-C9ORF002_ses-02_T1w.nii /media/mpasternak/ADATA-1TB/TMEM_Project/derivatives/spm/longitudinal/subjects/sub-C9ORF002/ses-11/mri/s10mwp1rsub-C9ORF002_ses-11_T1w.nii /media/mpasternak/ADATA-1TB/TMEM_Project/derivatives/spm/longitudinal/subjects/sub-C9ORF002/ses-12/mri/s10mwp1rsub-C9ORF002_ses-
Calls: read.LME.opts.batch ... dataTable.AFNI.parse -> dtCheck_overall -> dtCheck_img_exists -> system
Execution halted
Curiously the system call there seems to be broken off at the end with _ses- ...unless that's just done to save termnal space.
I will also attach the output of afni_system_check:
mpasternak@mpasternak /media/mpasternak/ADATA-1TB/TMEM_Project/derivatives/afni$ afni_system_check.py -check_all
-------------------------------- general ---------------------------------
architecture: 64bit ELF
cpu type: x86_64
system: Linux
release: 6.5.0-21-generic
version: #21~22.04.1-Ubuntu SMP PREEMPT_DYNAMIC Fri Feb 9 13:32:52 UTC 2
distribution: ubuntu 22.04 Jammy Jellyfish
number of CPUs: 24
apparent login shell: bash
shell RC file: .bashrc (exists)
--------------------- AFNI and related program tests ---------------------
which afni : /home/mpasternak/afni/abin/afni
afni version : Precompiled binary linux_ubuntu_16_64: Feb 22 2024
: AFNI_24.0.08 'Caracalla'
AFNI_version.txt : AFNI_24.0.08, linux_ubuntu_16_64, Feb 22 2024, official
which python : /usr/bin/python
python version : 3.10.12
which R : /usr/bin/R
R version : R version 4.3.2 (x86_64-pc-linux-gnu)
instances of various programs found in PATH:
afni : 1 (/home/mpasternak/afni/abin/afni)
R : 1 (/usr/bin/R)
python : 1 (/usr/bin/python3.10)
python2 : 1 (/usr/bin/python2.7)
python3 : 1 (/usr/bin/python3.10)
testing ability to start various programs...
afni : success
suma : success
3dSkullStrip : success
3dAllineate : success
3dRSFC : success
SurfMesh : success
3dClustSim : success
uber_subject.py : success
3dMVM : success
------------------------ dependent program tests -------------------------
checking for dependent programs...
which tcsh : /usr/bin/tcsh
which Xvfb : /usr/bin/Xvfb
checking for R packages...
rPkgsInstall -pkgs ALL -check : success
R RHOME : /usr/lib/R
------------------------------ python libs -------------------------------
++ module loaded: matplotlib.pyplot
module file : /usr/lib/python3/dist-packages/matplotlib/pyplot.py
matplotlib version : 3.5.1
++ module loaded: flask
module file : /usr/lib/python3/dist-packages/flask/__init__.py
flask version : 2.0.1
++ module loaded: flask_cors
module file : /usr/lib/python3/dist-packages/flask_cors/__init__.py
flask_cors version : 3.0.9
-------------------------------- env vars --------------------------------
PATH = /home/mpasternak/.local/share/pnpm:/home/mpasternak/.pyenv/plugins/pyenv-virtualenv/shims:/home/mpasternak/.pyenv/shims:/home/mpasternak/.pyenv/bin:/home/mpasternak/.local/bin:/home/mpasternak/MATLAB/R2020b/bin:/home/mpasternak/freesurfer/bin:/home/mpasternak/fsl/share/fsl/bin:/home/mpasternak/fsl/share/fsl/bin:/home/mpasternak/.cargo/bin:/home/mpasternak/.local/bin:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:/usr/games:/usr/local/games:/snap/bin:/snap/bin:/home/mpasternak/afni/abin
PYTHONPATH =
R_LIBS =
LD_LIBRARY_PATH =
DYLD_LIBRARY_PATH =
DYLD_FALLBACK_LIBRARY_PATH =
CONDA_SHLVL =
CONDA_DEFAULT_ENV =
----------------------------- eval dot files -----------------------------
----------- AFNI $HOME files -----------
.afnirc : found
.sumarc : found
.afni/help/all_progs.COMP : found
--------- shell startup files ----------
-- good: .tcshrc seems to contain 'source .cshrc'
-- considered operations: path, apsearch
-- note: followers should not need edits, so edit flags should be 0
(have 1 follower(s), which can be ignored)
dot file test : want 1 modifications across 3 files:
file path flatdir apsearch follower
--------------- ---- ------- -------- --------
.cshrc 0 0 1 0
.tcshrc 0 0 0 1
.bashrc 0 0 0 0
------------------------------ data checks -------------------------------
data dir : found AFNI_data6 under $HOME/afni (467851M Avail)
top history: ... [gangc]: redo 0.BGA as 1.bga and update run.MSS.txt
data dir : found AFNI_demos under $HOME/afni
top history: ...ct 2020 [taylorp]: updated scripts under FATCAT_DEMO
data dir : found suma_demo under $HOME/afni
top history: ...s_New/data/Build_tmp on Mon Mar 4 11:56:45 EST 2013
data dir : found afni_handouts under $HOME/afni
atlas : found TT_N27+tlrc under /home/mpasternak/afni/abin
------------------------------ OS specific -------------------------------
which apt-get : /usr/bin/apt-get
apt-get version : apt 2.4.11 (amd64)
have Ubuntu system: ubuntu 22.04 Jammy Jellyfish
have Ubuntu afni : Precompiled binary linux_ubuntu_16_64: Feb 22 2024
========================= summary, please fix: =========================
* just be aware: login shell 'bash', but our code examples use 'tcsh'
* dot file test : want 1 modifications across 3 files:
I wonder if the dot file has something to do with this. I checked an earlier thread where another user had dot file test issues, so here is some more info to help you help me:
mpasternak@mpasternak /media/mpasternak/ADATA-1TB/TMEM_Project/derivatives/afni$ which afni
/home/mpasternak/afni/abin/afni
mpasternak@mpasternak /media/mpasternak/ADATA-1TB/TMEM_Project/derivatives/afni$ which init_user_dotfiles.py
/home/mpasternak/afni/abin/init_user_dotfiles.py
mpasternak@mpasternak /media/mpasternak/ADATA-1TB/TMEM_Project/derivatives/afni$ init_user_dotfiles.py -test -shell_list tcsh sh -do_updates path apsearch -verb 2
-- processing...
++ have original abin /home/mpasternak/afni/abin
-- setting dir_bin to ABIN /home/mpasternak/afni/abin
-- setting dir_dot to $HOME
++ .cshrc : 11 lines
++ .tcshrc : 3 lines
++ .profile : 38 lines
-- good: .tcshrc seems to contain 'source .cshrc'
-- skipping apsearch file shell tcsh: 1
-- skipping apsearch file shell tcsh: 1
-- skipping apsearch file shell sh: 1
-- considered operations: path, apsearch
-- note: followers should not need edits, so edit flags should be 0
(have 1 follower(s), which can be ignored)
dot file test : want 1 modifications across 3 files:
file path flatdir apsearch follower
--------------- ---- ------- -------- --------
.cshrc 0 0 0 0
.tcshrc 0 0 0 1
.profile 1 0 0 0
mpasternak@mpasternak /media/mpasternak/ADATA-1TB/TMEM_Project/derivatives/afni$ init_user_dotfiles.py -ver
init_user_dotfiles.py, version 1.5, January 2, 2024
For the record, the 2nd lattermost command doesn't seem to actually make any changes...should it?
It's refreshing that you guys have actual LMEr capability, so please let me know if I can provide anything to help troubleshoot this.